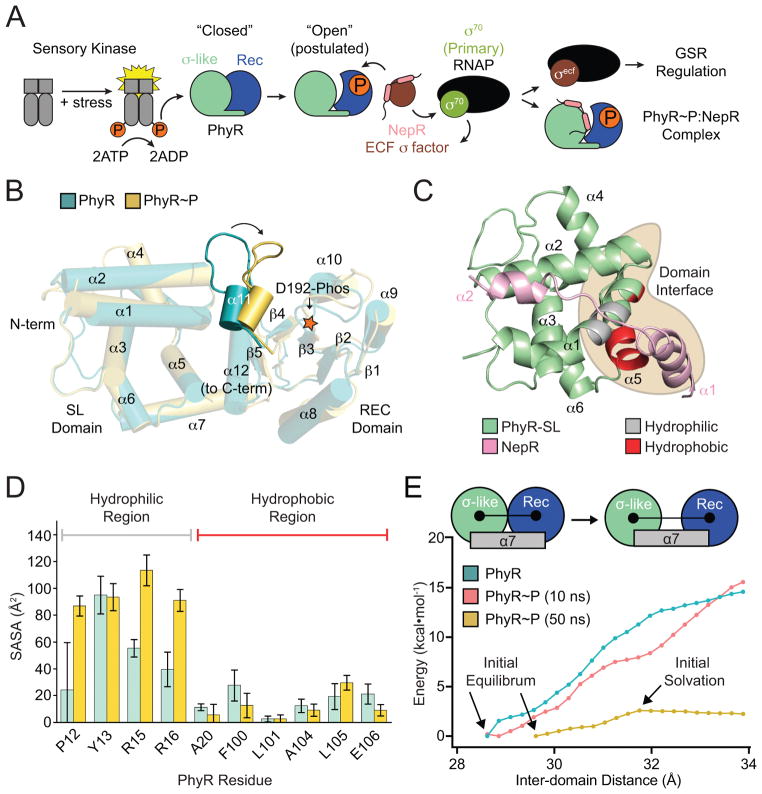
Figure 1. PhyR α11-β5 loop rearrangement decreases the energetic barrier to opening.
(A) A model of general stress response (GSR) activation. Upstream sensory kinases phosphorylate PhyR, leading to PhyR opening and NepR binding. This releases σEcfG to direct GSR transcription. (B) Alignment of PhyR (green) and PhyR~P (gold) MD trajectories. Rearrangement can occur on short MD timescales in the α11-β5 loop, which retracts toward the REC domain (shown by arrow). Structures shown are the average backbone positions over the final 10 ns of simulation. (C) NepR (pink) modelled into the PhyR-SL domain (green) binding site using a hypothetical open state model. Hydrophobic (red) and hydrophilic (gray) regions at the domain interface (brown) are highlighted. (D) Calculated solvent accessible surface area (SASA) values for PhyR (green) and PhyR~P (gold) at 50 ns for NepR-binding residues. (E) Umbrella sampling of PhyR and PhyR~P at increasing inter-domain distances. Arrows indicate points corresponding to the initial equilibrium state of the protein, collected from previous simulations, as well as the point in the PhyR~P (50 ns) sampling where solvation of the total interdomain interface began. The energetic barrier to open PhyR is lowered after phosphorylation (see 50 ns simulation). Energy to open PhyR~P prior to solvation is equivalent to unphosphorylated PhyR. Error bars represent standard deviation.