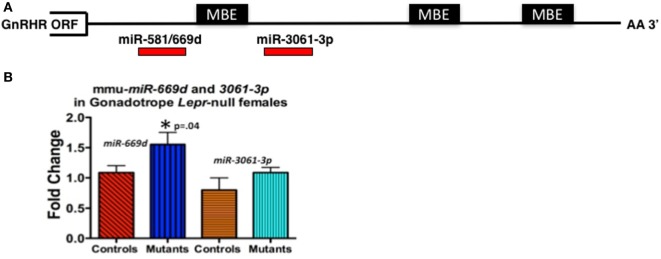
Figure 4.
(A) Candidate regulatory elements within the murine Gnrhr 3′-untranslated region (UTR) include MBEs and at least two miRNA target sites. In silico analyses (TargetScan 7.1, ENSMUST00000031172.8) indicate three consensus MSI binding sites or elements (MBEs) and 16 miRNAs target sites (only 2 shown) within the murine GnRHR 3′UTR (182 nucleotides). The relative positions of the three MBEs and two miRNAs (miR-581/669d and miR-3061-3p) are shown schematically. The MBE closest to the open-reading frame (ORF) is flanked by sequences encoding miR-581/669d and miR-3061-3p. (B) miR-581/669d and miR-3061-3p levels in Lepr-null gonadotropes. Total RNA enriched for miRNA was isolated from whole pituitaries of control and gonadotrope-LeprEx17-null females using the Maxwell miRNA tissue kit (Promega, AS1470). We used the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, 4366596) with TaqMan small RNA assays to amplify our miRNAs of interest. We performed qRT-PCR using the TaqMan small RNA qPCR primers with the TaqMan Universal PCR Master Mix II (Applied Biosystems, 4440038) in triplicate. We used the protocol provided with the master mix and performed the experiment using a QuantStudio 12k Flex (Applied Biosystems) and the protocol provided with the master mix. Real-time PCR showed miR-581/669d elevated in the absence of leptin signaling, which is consistent with a role for this miRNA as a repressor of Gnrhr mRNA translation. *Significantly different from controls by Student’s t-test.