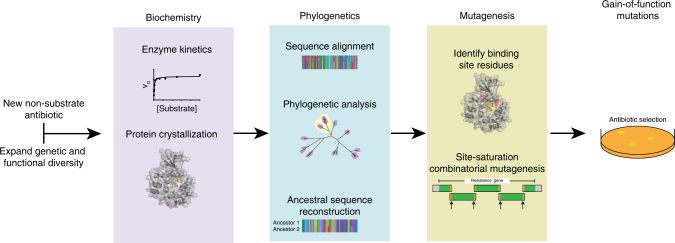
Fig. 7.
Workflow for assessing the evolvability of resistance enzymes to new antibiotics using biochemistry, phylogenetics, and mutagenesis. First, expand the known functional diversity of an enzyme class by identifying homologs that include the new antibiotic in its substrate range. Characterize the enzymes using steady-state kinetics and solve the protein structure of at least one homolog. Use the biochemical experiments to correlate functional diversity with phylogenetic diversity for identifying the last common ancestor of each functional subtype (e.g., ancestor of narrow range Mphs and wide range Mphs, respectively). The difference in ancestral protein sequences are residues possibly involved in substrate specificity, which should be prioritized based on proximity to the substrate-binding site in the protein structure. Target these residues for mutagenesis, preferably by saturation mutagenesis at each position, in combination (site-saturation combinatorial mutagenesis). Identify residues expanding substrate specificity by sequencing technologies that generate reads covering the whole gene (e.g., Sanger, Nanopore, PacBio)