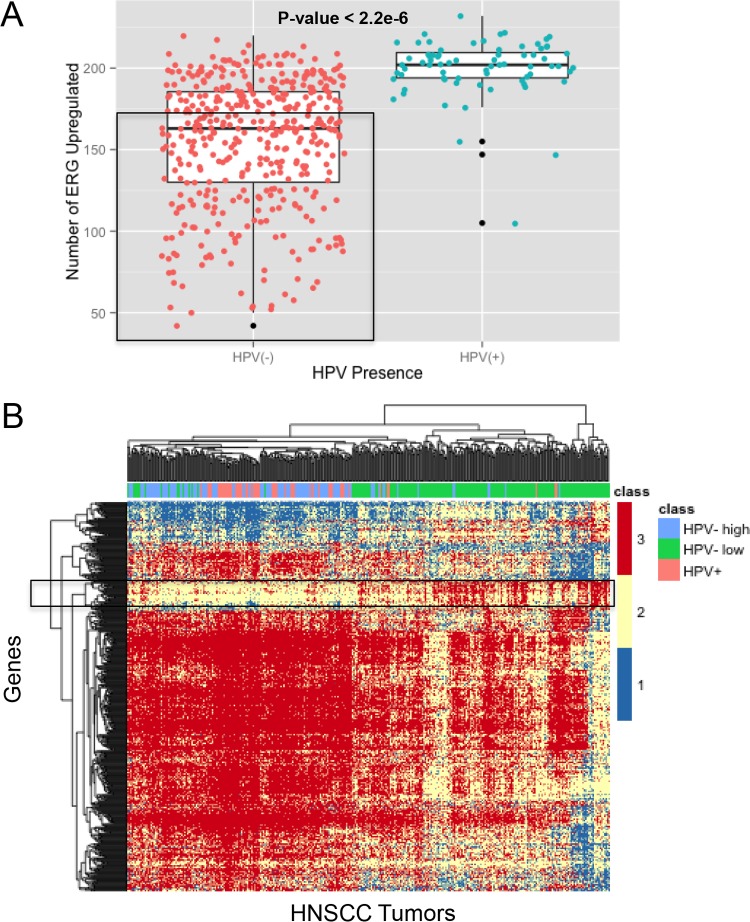
FIG 2 .
HNSCC tumors partition into three groups based on E2F-regulated gene expression. (A) Gene expression in HNSCC tumors was categorized as described in Materials and Methods. The total number of genes upregulated out of the 325 E2F-regulated genes (Table S2 in Data Set S1) was quantified for each patient. HPV− patient data are marked by red circles, HPV+ patient data by blue circles, and outliers by black circles. Three groups are defined based on the number of ERGs upregulated and the presence of HPV: HPV+, high-ERG tumors; HPV−, high-ERG tumors; and HPV−, low-ERG tumors (the latter group is bounded by the rectangle). The t test was used to compare the difference between the results for HPV+ and HPV− samples. (B) Analysis of ERG-defined HNSCC groups shows a subset of genes uniquely upregulated in the HPV−, low-ERG group. The global mRNA levels in the three HNSCC groups were analyzed. Gene expression was categorized with a cutoff of 1 (see Materials and Methods), and an ANOVA/Tukey test was run to find genes differentially regulated between the HPV−, low-ERG group and HPV− or HPV+, high-ERG tumors. The resulting 586 genes were clustered, and the results displayed as a heatmap. Genes that are uniquely upregulated in HPV−, low-ERG tumors are highlighted with a rectangle.