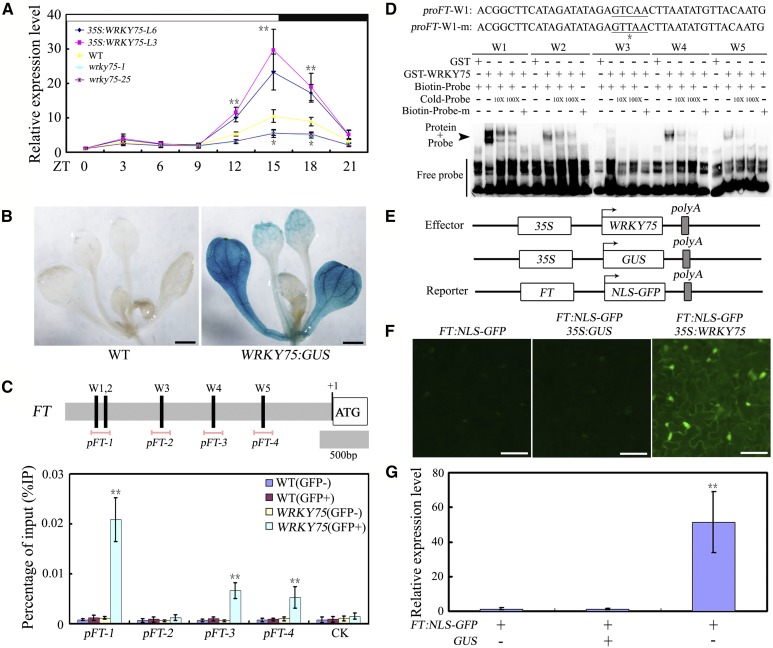
Figure 3.
WRKY75 promotes flowering by activating FT transcription. A, Expression of FT in the indicated genotypes. Ten-d-old plants grown under normal growth conditions (22°C, LD) were harvested at the indicated ZT for total RNA extraction and qRT-PCR assays. Transcript levels of FT in untreated Col-0 leaves at ZT0 were arbitrarily set to 1. Values are mean ± sd of three independent biological replicates. Two-way ANOVA was performed for statistical analysis; asterisks indicate significant differences as compared to controls, *P < 0.05, **P < 0.01. B, WRKY75:GUS expression in 12-d-old seedlings. The expression of WRKY75 was detected by GUS staining. Representative seedlings were photographed. Scale bar: 1 mm. C, The promoter structure of the FT gene and fragment used in the ChIP assay. The upper panel shows schematic representation of the FT promoter regions containing W-box clusters. The diagram indicates the number and relative position of the W-boxes in the respective promoters relative to the ATG start codon. In the promoter fragment names, the prefix “p” indicates promoter. Pink lines indicate the sequences detected by ChIP assays. ChIP assays were performed with chromatin prepared from WRKY75:YFP-WRKY75:3′-WRKY75 transgenic plants, using an anti-GFP antibody (immunoprecipitated). ChIP results are presented as a percentage of input DNA. Values are mean ± sd of three independent biological replicates. Asterisks indicate Student’s t-test significant differences as compared to controls, **P < 0.01. D, The EMSA analysis of the binding of recombinant WRKY75 protein to the promoter of FT. The oligonucleotides (proFT-W1/2/3/4/5 and proFT-W1/2/3/4/5-m) were used as the probes. Underlining signifies W-box sequence and asterisk represents the mutated base in the W-box element (as exampled with W1). GST, GST-WRKY75, biotin-probe, labeled mutated probe, and unlabeled probe at a 10× and 100× molar excess were present (+) or absent (−) in each reaction. E, Schematic of the FT:NLS-GFP reporter and WRKY75 and GUS effectors. F, Transient expression assays showed that WRKY75 activates the expression of FT. GFP fluorescence was detected 48 h after coinfiltration with the indicated constructs. The experiment was repeated three times with similar results and representative photos were displayed. Scale bar, 50 μm. G, qRT-PCR analysis of the accumulation of GFP transcripts. Total RNAs were extracted from leaves of N. benthamiana coinfiltrated with combinations of various constructs in (E). The N. benthamiana ACTIN gene was used as an internal control. Values are mean ± sd of three independent biological replicates. Asterisks indicate Student’s t-test significant differences as compared to controls, **P < 0.01. Biotin-Probe-m, biotin-labeled probe with a single nucleic acid mutation from TGAC to TAAC; Cold-Probe, unlabeled probe.