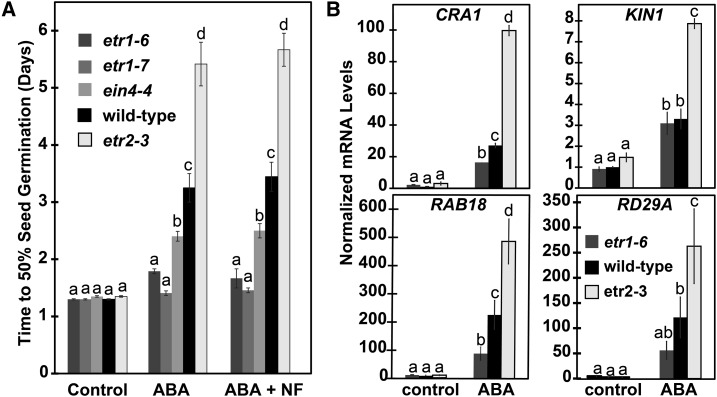
Figure 3.
ETR1 and EIN4 affect responses to ABA oppositely from ETR2. A, Germination time-courses of wild-type, etr1-6, etr1-7, ein4-4, and etr2-3 seeds in response to 1 μM ABA and 1 μM ABA plus 100 μM NF to block ABA biosynthesis were conducted and the times for 50% seed germination were determined. Data represents the average ± sd. B, The transcript abundance of RAB18, CRA1, KIN1, and RD29A were measured in wild-type, etr1-6, and etr2-3 seeds using real-time qRT-PCR. Seeds were germinated for 2 d in the presence or absence of 1 μM ABA and mRNA extracted. Data were normalized to the levels of At3g12210 in each seed line to calculate the relative transcript level for each gene. These were then normalized to levels of the transcript in untreated wild-type seeds. The average ± se for two biological replicates with three technical replicates each is shown. For both panels, 0.01% (v/v) ethanol was used as a solvent control. Data were analyzed using a two-way ANOVA and Tukey’s multiple comparisons test. In each panel, the different letters indicate significant difference (P < 0.05).