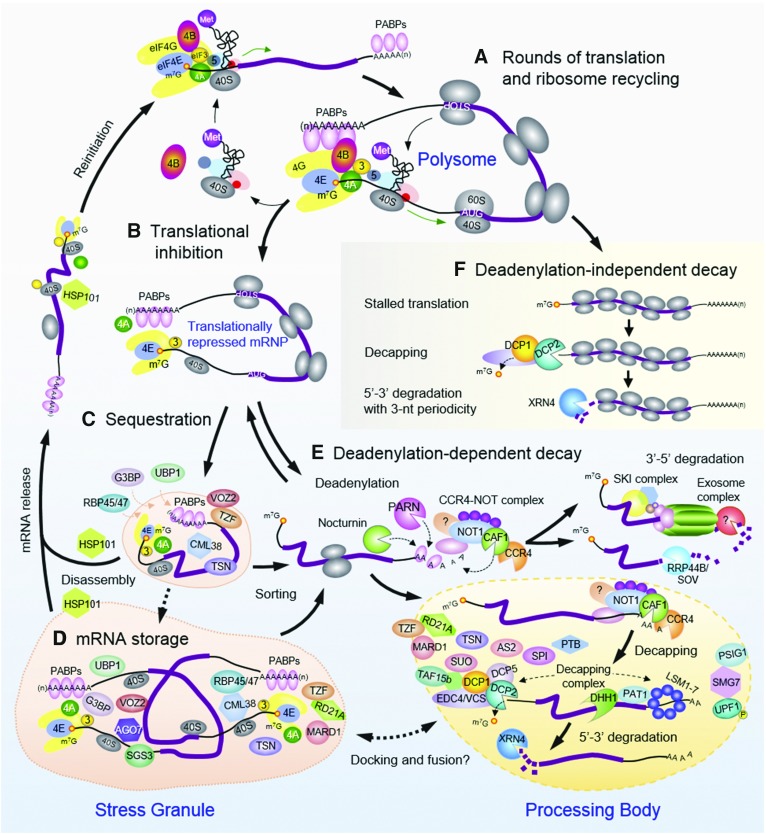
Figure 2.
Overview of cytoplasmic mRNA translation, storage, and decay in plants. A, Cytoplasmic mRNAs undergoing active translation form polysomes and remain in a translationally active state until damaged or translationally repressed. B, Repression can result from multiple causes (i.e. altered abundance or phosphorylation of specific translation factors or RNA-binding proteins and ribosome stalling) that limit translational initiation or ribosome translocation, promoting transition into a translationally repressed mRNP, where the mRNA is either sequestered by RNA-binding proteins (e.g. UBP1, G3BP, and RBP45/47) or degraded via different pathways. C, mRNA sequestration is typically triggered by cellular stress, serving as a sorting nucleation point for mRNP assembly into SGs for storage or PBs for degradation. D, During a stress-recovery period, intact mRNAs stored in SGs can reenter translation via a process facilitated by a chaperone (e.g. HSP101 during heat stress recovery). Some mRNAs released from SGs may be targeted for degradation after stress. E, In general mRNA decay, the 3′ protective poly(A) tail is removed by different classes of deadenylases [Nocturnin, poly(A)-specific RNase (PARN), and the CCR-NOT complex], then degradation proceeds in the 3′ to 5′ direction by the multimeric SKI-exosome complex and/or the RRP44B/SOV exonuclease. After deadenylation, mRNA degradation also can occur from the 5′ to 3′ direction. mRNAs degraded via this mode could be localized in the cytosol and/or PBs, where the multiprotein mRNA-decapping complex removes the protective 5′-cap and, subsequently, the 5′-3′ XRN4 exoribonuclease catalyzes nucleotide hydrolysis. F, In addition to the deadenylation-dependent decay pathway, translationally repressed mRNAs can be degraded directly in the 5′ to 3′ direction while in association with elongating ribosomes via XRN4-mediated cotranslational decay. This mode of degradation bypasses deadenylation but may require mRNA decapping. The progressive 5′-3′ exonucleolytic destruction of the mRNA by XRN4 yields a codon-by-codon three-nucleotide signature that is thought to reflect the movement of the most 5′ ribosome along the transcript. AUG, Translation initiation codon; STOP, termination codon.