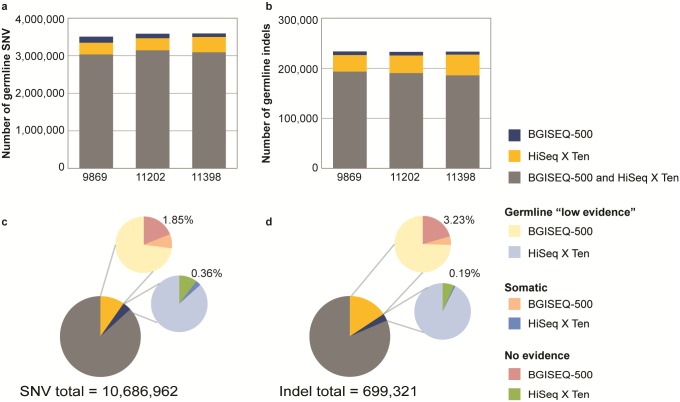
Fig 2. Germline variants identified in three mesothelioma samples (patients: 9869, 11202 and 11398) using BGISEQ-500 and HiSeq X Ten data.
The number of germline SNV (a) and indels (b) identified in each patient using the BGISEQ-500 and HiSeq X Ten platforms. We investigated germline SNV (c) and indels (d) which were only called in one platform and that fall into three categories: i) identified as germline in the other platform but with low evidence; ii) identified in the other platform but predicted as a somatic variant; or iii) not identified in the other platform. Across the 3 patients only 197,434 (1.85%) SNVs were truly unique to the HiSeq X Ten and not identified in the BGISEQ-500 (c). Similarly in the BGISEQ-500 platform only 38,236 SNVs (0.36% of the total) were truly unique to the BGISEQ-500, not called in the HiSeq X Ten data (c). The same pattern was observed for indels (d), only 3.23% were unique to HiSeq X Ten and 0.19% to BGISEQ-500.