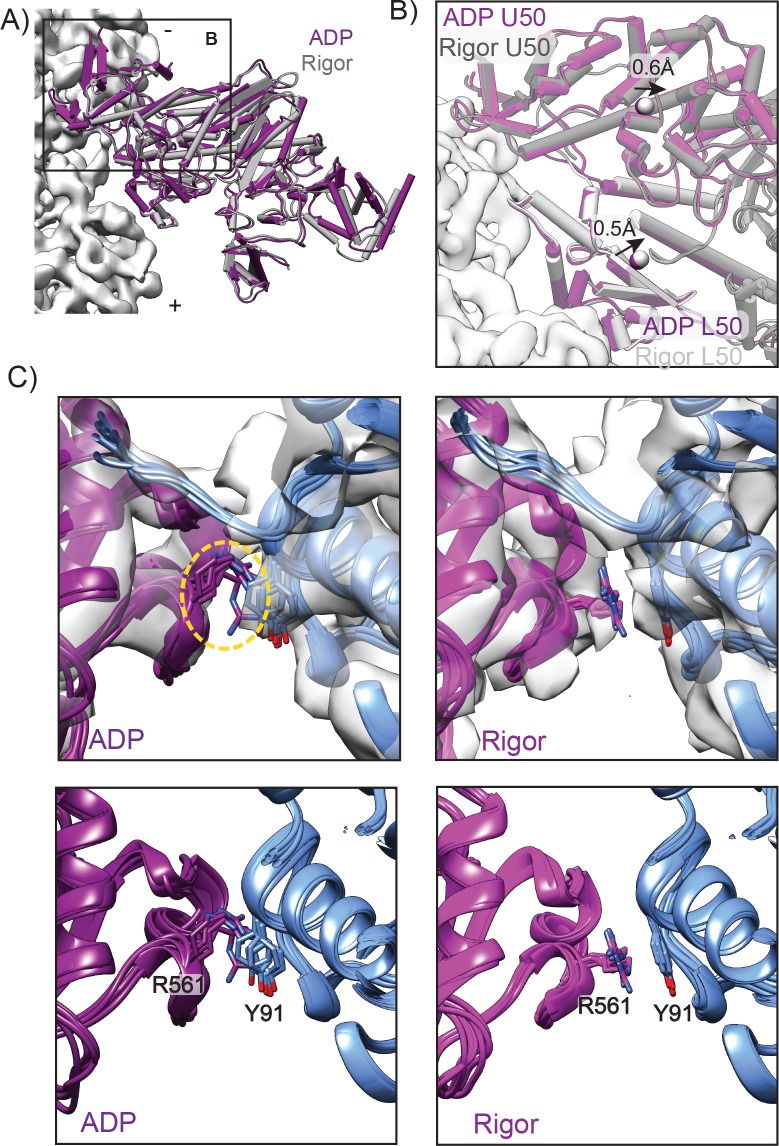
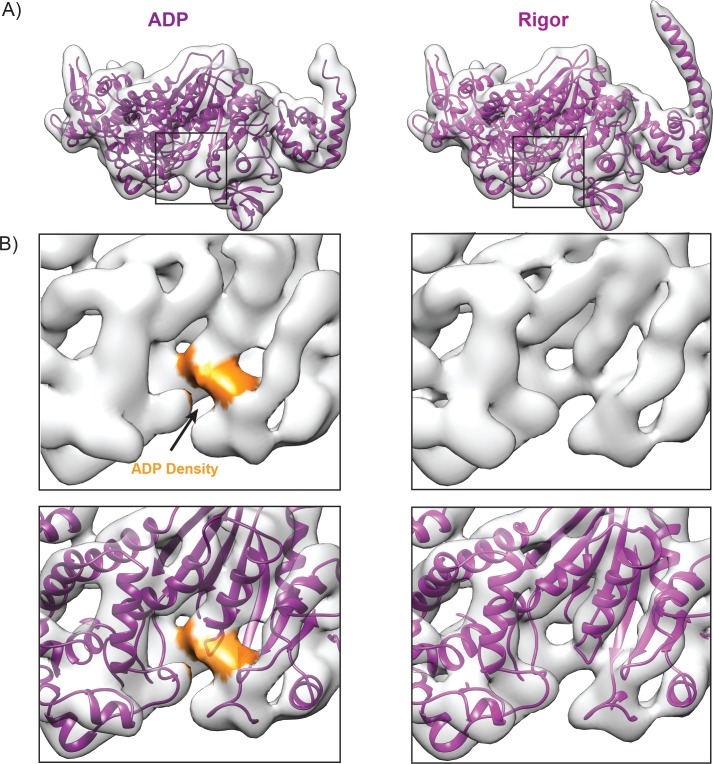
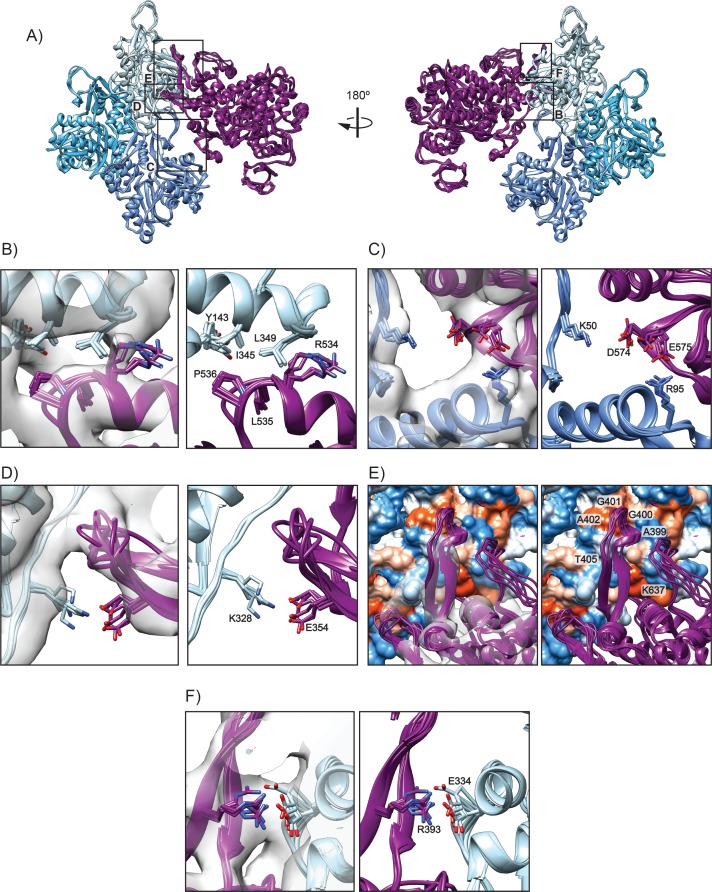
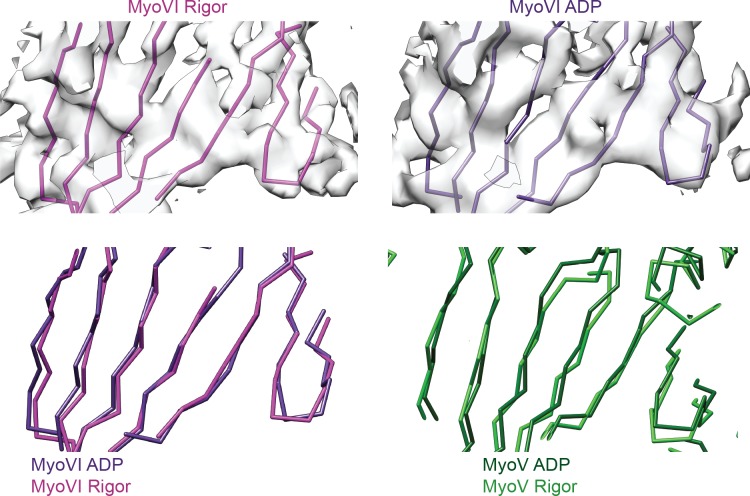
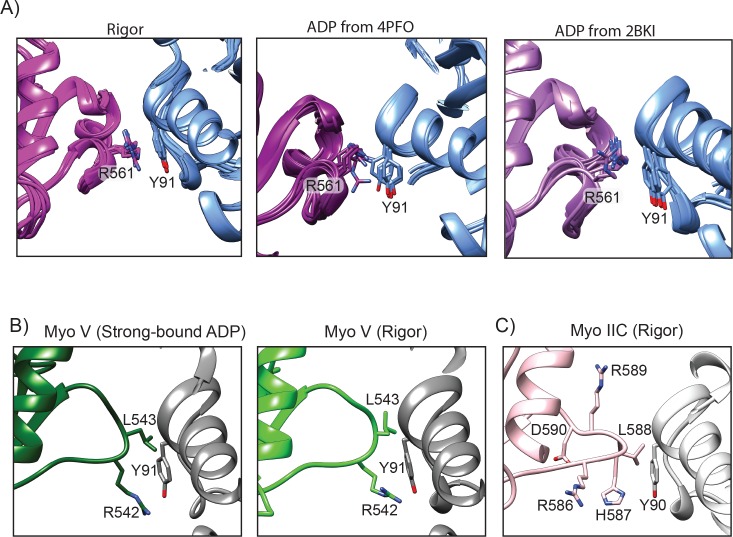
Figure 3. A unique contact is established upon transition from ADP to rigor.
(A) View of the LPF APD MDFF model (dark magenta) and LPF rigor MDFF model superimposed in the reference frame of the actin filament (light gray density). To generate this superposition, the ADP and rigor density maps were aligned, then their corresponding atomistic models were rigid body fit into the aligned maps. (B) Minimal actin binding cleft rearrangements are observed between ADP and rigor, superimposed as described in A. ADP U50, magenta; ADP L50, dark magenta; rigor U50, dark grey; rigor L50, light grey; actin density, white. Arrows denote displacements of domain centroids (spheres) from ADP to the rigor state. Centroids were determined for U50 (residues 180–206, 229–397, and 405–441) and L50 (residues 467–597 and 638–661) domains. (C) MDFF indicates the Milligan contact cation-π interaction between R561 in the MD loop 3 and Y91 in the adjacent actin is absent in ADP (left) but is established upon transition to the rigor state (right), with clear density for these sidechains in the rigor map. For both states, all six actomyosin interfaces in the corresponding HR MDFF model are displayed superimposed on one actin subunit as described in Figure 2A. Density maps are displayed in transparent grey in the upper panels. Orange dotted circle indicates absence of density for R561 in the ADP map, while density for Y91 is still present.