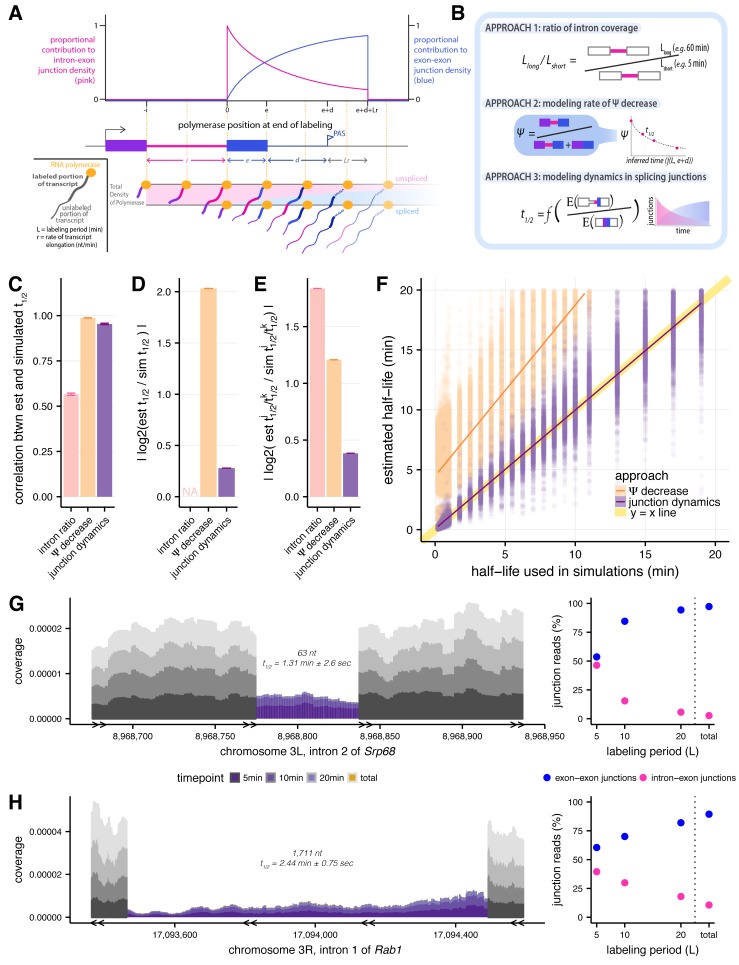
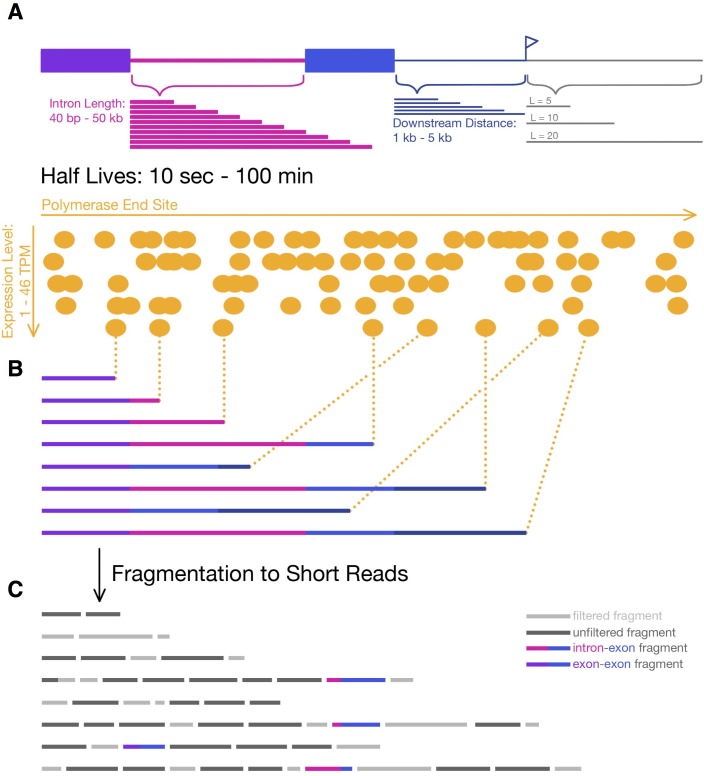
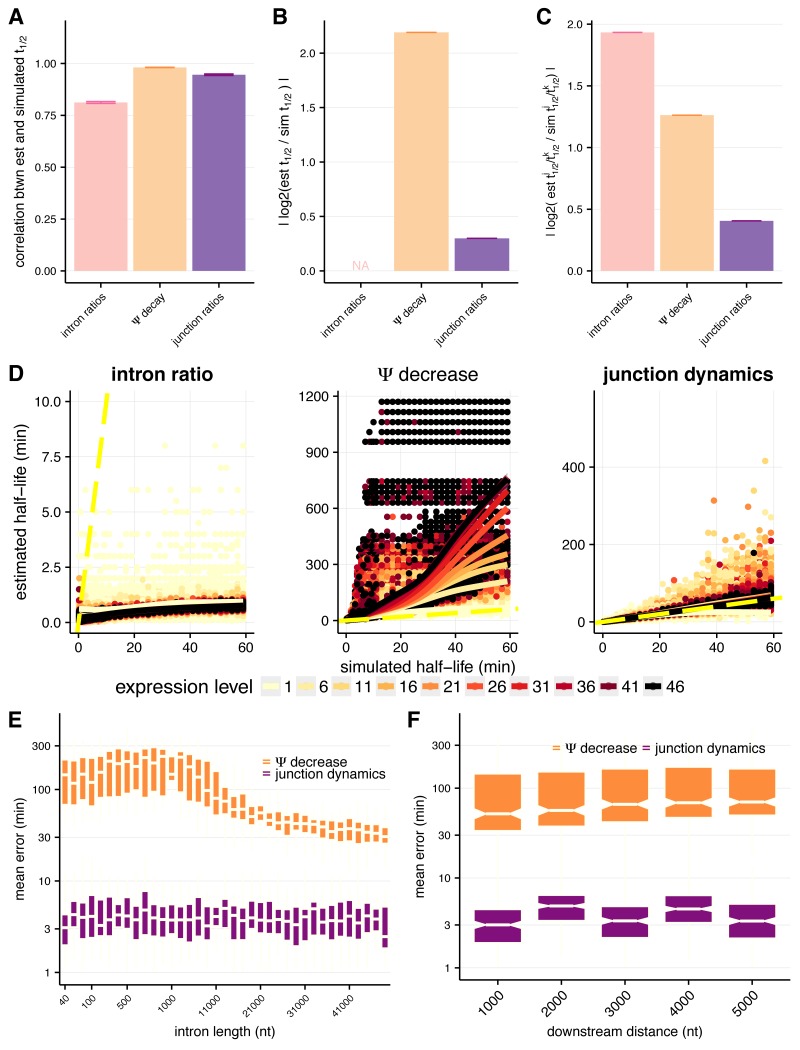
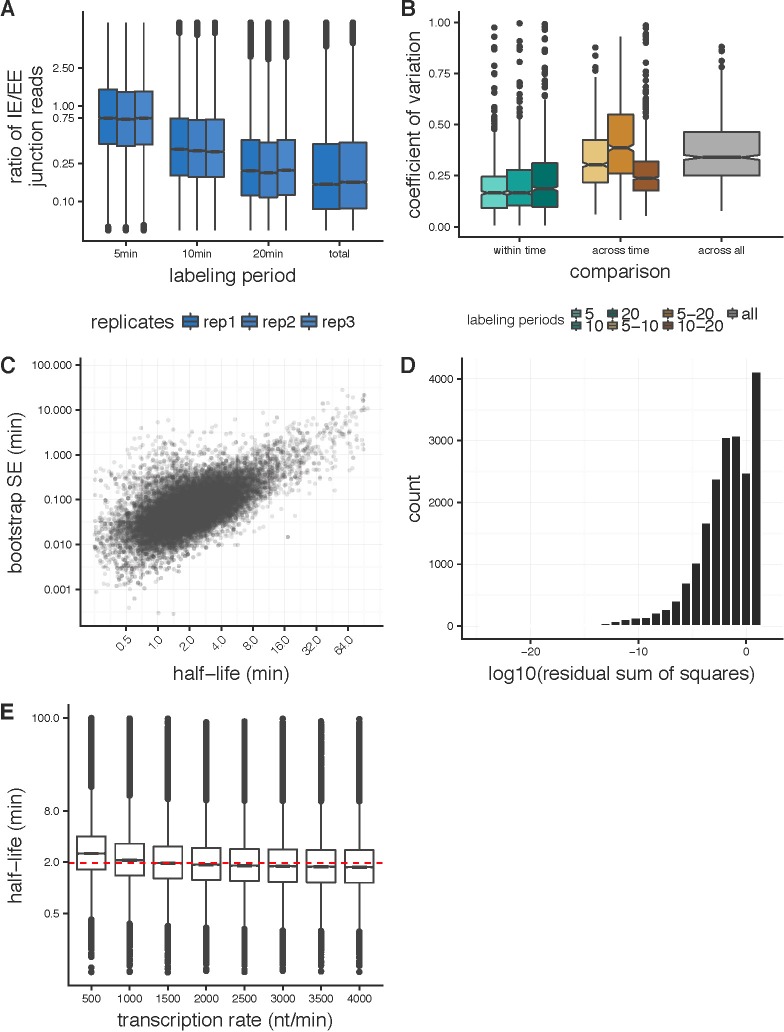
Figure 1. Estimating intron-specific splicing half-lives.
(A) Progressive labeling with 4sU results in sampling of nascent RNA molecules from polymerase molecules distributed across a gene (bottom). The probability of sampling reads from unspliced or spliced transcripts, represented by intron-exon (pink) and exon-exon (blue) junction reads respectively, is dependent on the intron half-life and the location of the polymerase at the completion of the labeling period (top). (B) Schematics outlining the three approaches assessed for measuring rates of intron excision. (C) Mean Spearman correlations between simulated half-lives and estimated splicing rates from each of our three approaches (error bars are ± standard error). (D) Absolute percent error of our estimated splicing rates relative to simulated half-lives. Intron ratios are relative measures of half-lives, thus were not included in this comparison (error bars are ± standard error). (E) Relative absolute percent error of the estimated and simulated half-lives between two introns, allowing comparisons of metrics not expected to be drawn from the same distribution (error bars are ± standard error). (F) Estimated half-lives from Ψ decrease and junction dyanmics approaches (x-axis) versus to the half-lives used to simulate read data (y-axis). Yellow line indicates y = x line of perfect correlation. (G and H) Nascent RNA coverage across the second intron of Srp68 (G) and the first intron of Rab1 (H). Colors represent time points, with 5 min after 4sU labeling (darkest shade), through 10 min, 20 min, and total RNA sample (lightest shade). Right panels show the proportions of intron-exon (pink) and exon-exon (blue) junction reads out of all 3’ junction reads in each labeling period.