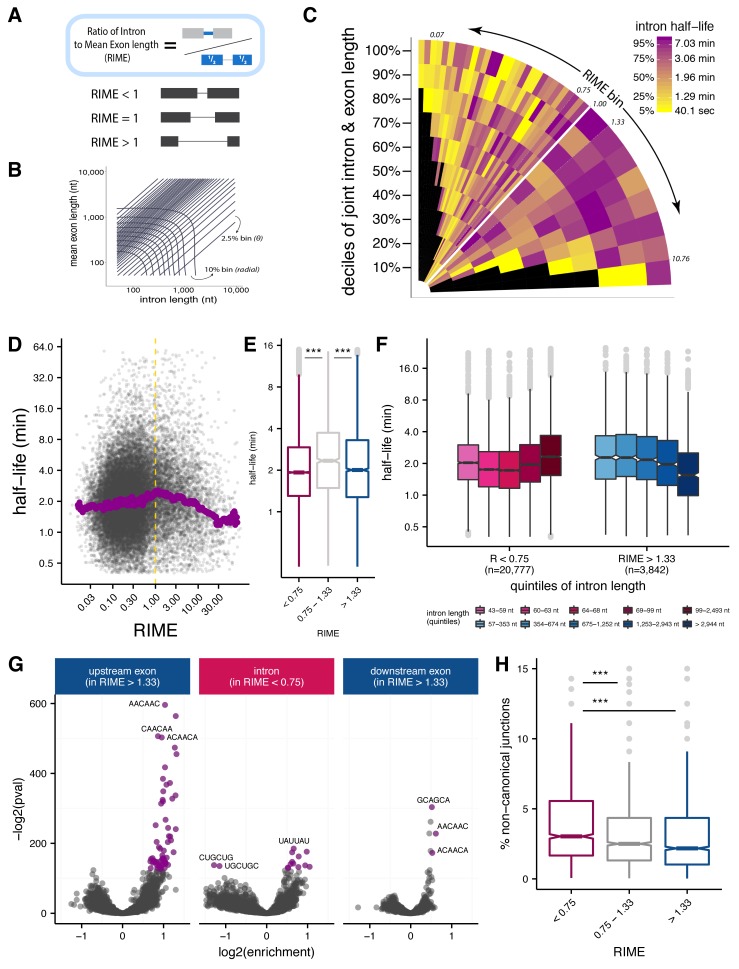
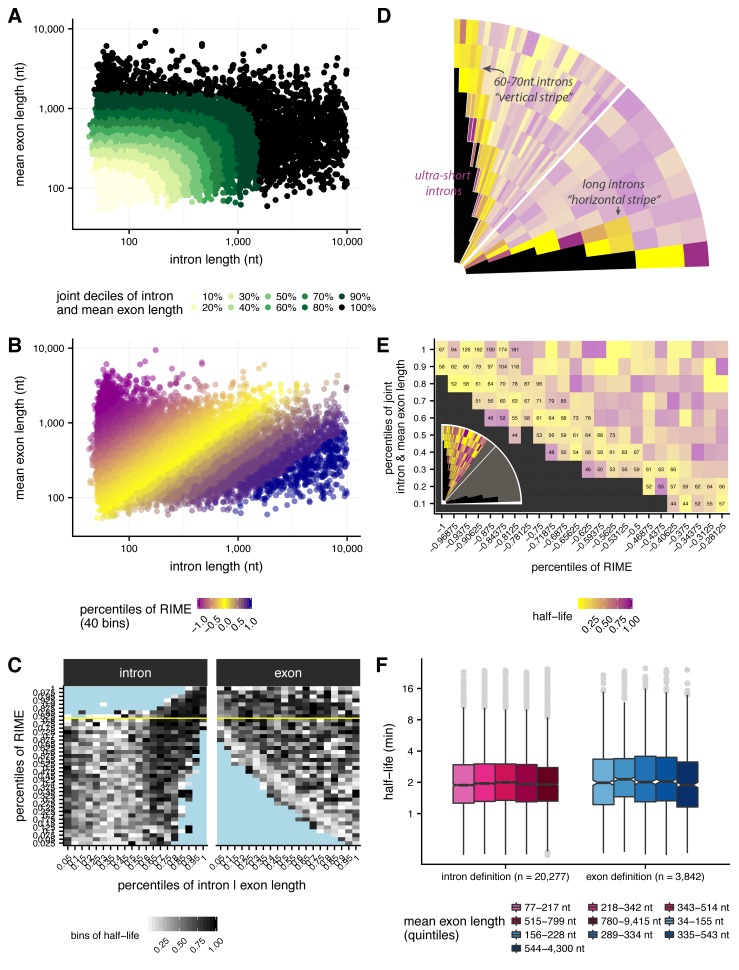
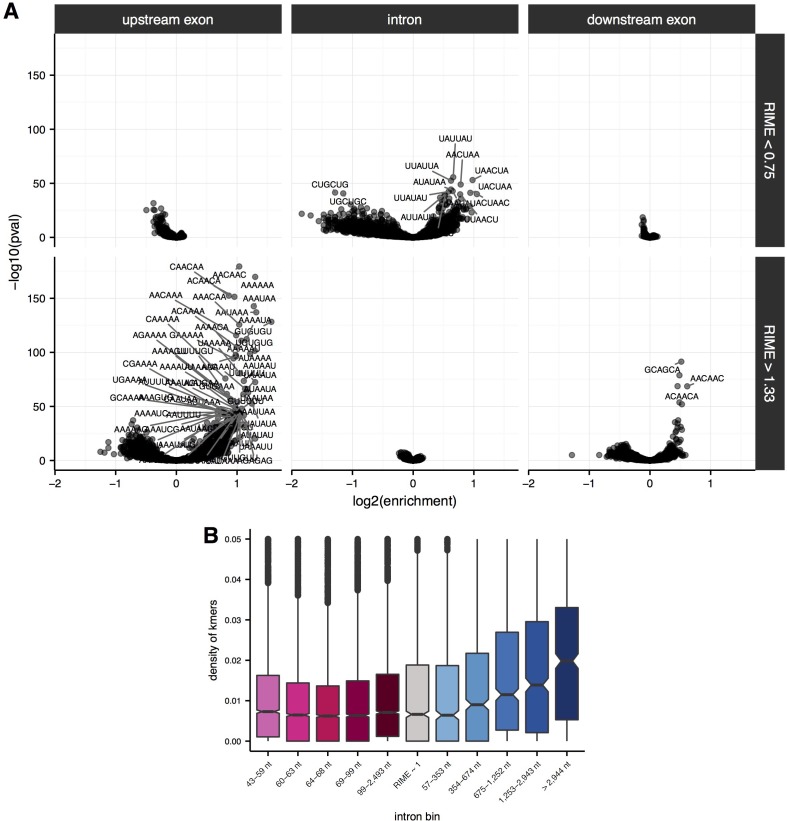
Figure 3. Splice site recognition mode influences the efficiency of splicing.
(A) The Ratio of Intron to Mean Exon (RIME) metric is defined as the ratio of a given intron length to the mean length of the exons flanking that intron. (B) Schematic of binning to capture intron length (x-axis, nt), mean flanking exon length (y-axis, nt), and RIME values in a single plot. Radial bins capture both intron and and mean exon length (10% bins), while diagonal bins (2.5% bins, θ axis) capture the RIME distribution. (C) Distribution of mean splicing half-lives across bins of RIME (θ axis) and deciles of joint intron and exon lengths (r axis). Yellow represents short mean half-lives and dark purple represents longer mean half-lives. (D) Running median of splicing half-lives across distribution of RIME values. Median is computed in sliding bins of 200 introns. (E) Distribution of splicing half-lives for introns with RIME <0.75 (pink), 0.75 < RIME < 1.33 (grey), and RIME >1.33 (blue). (F) Splicing half-lives across quintiles of intron length in each RIME class (RIME <0.75 in shades of pink on left and RIME >1.33 in shades of blue on right). (G) Enrichment of 6mers in exons upstream of introns with RIME >1.33 (left), intronic regions of introns with RIME <0.75 (middle), and exon downstream of introns with RIME >1.33 (right). Significant 6mers are in purple (Benjamini-Hochberg corrected p-value<10−30). (H) Splicing accuracy measured by percentage of non-canonical unannotated reads for introns with RIME <0.75 (pink), 0.75 < RIME < 1.33 (grey), and RIME >1.33 (blue).