Figure 1.
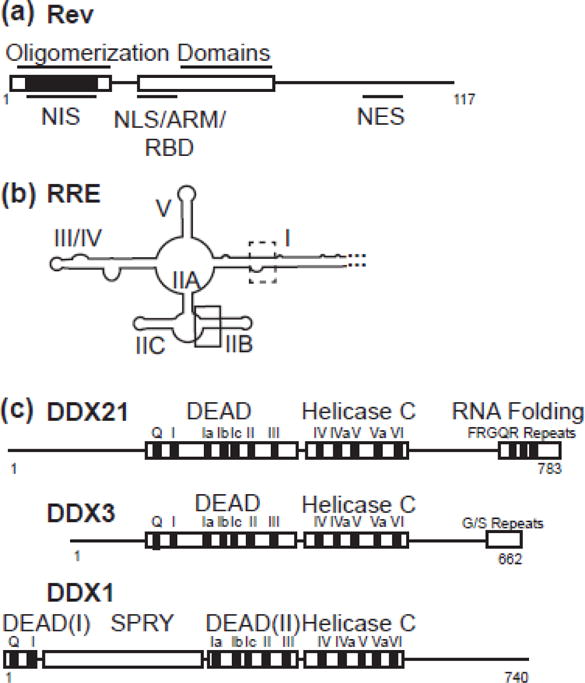
Schematic diagrams of proteins and RNA used in this study. a) Rev, with structured helix-turn helix domains boxed. Oligomerization domains, nuclear export inhibitory signal (nis), nuclear localization signal (NLS), arginine rich motif (ARM), RNA binding domain (RBD) and nuclear export signal (NES) labelled. NIS region has additionally been colored black. b) RRE RNA molecule, with stems I, IIA, IIB, IIC, III/IV, and V labelled. High affinity primary binding site boxed in black, and lower affinity binding site boxed in dotted line. c) DDX21, DDX3 and DDX1 diagrams aligned to DEAD domain and Helicase C domain interface. Accessory domains are labelled.
