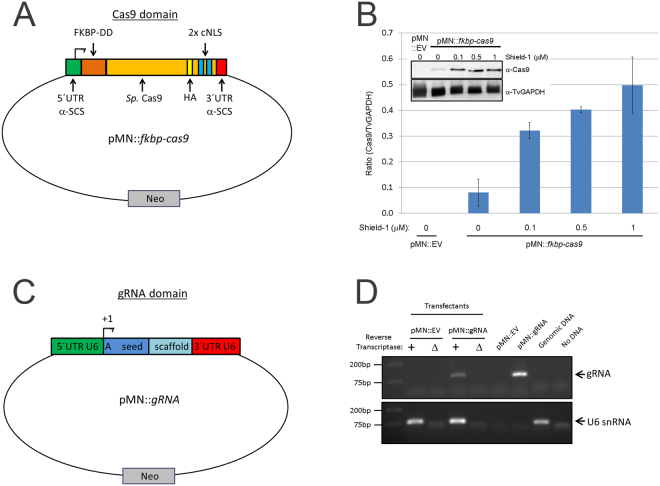
Figure 1.
Setup of pMN::fkbp-cas9 and pMN::gRNA constructs. (A) Cartoon representation (not to scale) of pMN::fkbp-cas9 plasmid. The T. vaginalis gene α-SCS 5′ and 3′ UTRs of the pMasterNEO plasmid (pMN)53 drive expression of the fkbp(FKBP destabilization domain)-cas9 fusion gene. The cas9 variant used is the human codon optimized version with 2 SV40 nuclear localization signals at the C-terminus56. (B) Representative immunoblot analysis (inset) and quantification of replicate samples of pMN::fkbp-cas9 expression. Samples were induced for 24 hours with 0, 0.1, 0.5 or 1 μM Shield-1 ligand before protein was collected and resolved by SDS-PAGE. Inset, upper: anti-Cas9 immunoblot (Clontech); Inset, lower: anti-TvGAPDH antibody (Cocalico Biologicals). Control sample used a pMN::EV (pMasterNEO::empty vector) transfectant. Quantitative comparison of samples utilized normalized signal of Cas9 (upper blot) to GAPDH signal (lower blot) taken from within an experiment and analyzed in parallel on separate immunoblots using a Bio-Rad Gel Doc and ImageLab software. Bar graph represents average +/− standard deviation of two independent analyses. (C) Cartoon representation (not to scale) of pMN::gRNA – 360 bp of the T. vaginalis U6 5′ UTR and 37 bp of the 3′ UTR flank the gRNA. A 20 nt seed region with the first nucleotide always an adenine residue followed by 19 nt seed region used for specific targeting and the gRNA scaffold. (D) RT-PCR products amplifying either the gRNA sequence or the U6 snRNA (control). Total RNA was subjected to +/− reverse transcriptase then amplified by PCR using gRNA- or U6 snRNA-specific primers. Control PCRs included the pMN::EV and pMN::gRNA plasmids, genomic DNA and no DNA template. PCR products were imaged using a Bio-Rad Gel Doc and ImageLab software. Full length blots/gels are presented in Supplementary Figure S5.