Abstract
Phosphorylation of the eukaryotic translation initiation factor, eIF2α, by stress-activated protein kinases and dephosphorylation by the growth arrest and DNA damage-inducible protein (GADD34)-containing phosphatase is a central node in the integrated stress response. Mass spectrometry demonstrated GADD34 acetylation at multiple lysines. Substituting K315 and K322 with alanines or glutamines did not impair GADD34’s ability to recruit protein phosphatase 1α (PP1α) or eIF2α, suggesting that GADD34 acetylation did not modulate eIF2α phosphatase activity. Arsenite (Ars)-induced oxidative stress increased cellular GADD34 levels and enhanced Sirtuin 1 (SIRT1) recruitment to assemble a cytoplasmic complex containing GADD34, PP1α, eIF2α and SIRT1. Induction of GADD34 in WT MEFs paralleled the dephosphorylation of eIF2α (phosphoserine-51) and SIRT1 (phosphoserine-47). By comparison, eIF2α and SIRT1 were persistently phosphorylated in Ars-treated GADD34−/− MEFs. Expressing WT GADD34, but not a mutant unable to bind PP1α in GADD34−/− MEFs restored both eIF2α and SIRT1 dephosphorylation. SIRT1 dephosphorylation increased its deacetylase activity, measured in vitro and in cells. Loss of function of GADD34 or SIRT1 enhanced cellular p-eIF2α levels and attenuated cell death following Ars exposure. These results highlighted a novel role for the GADD34/PP1α complex in coordinating the dephosphorylation and reactivation of eIF2α and SIRT1 to determine cell fate following oxidative stress.
Aberrations in protein homeostasis (proteostasis), particularly the processing and folding of proteins in the endoplasmic reticulum (ER), creates an ‘ER stress’ that is a contributing factor in many chronic or aging-related human diseases, including cancer, diabetes, inflammatory or heart disease and neurodegeneration.1 The ER, characterized by its unique oxidizing environment, is well suited for the formation of disulphide bonds, promoting the folding of proteins into their three-dimensional state. The accumulation of misfolded proteins that creates ER stress activates the ER-transmembrane sensors, PERK, ATF6 and IRE1, to control protein synthesis, folding and degradation and restore proteostasis. The failure to establish normal protein folding, an energy demanding process, creates an oxidative stress with reactive oxygen species (ROS) production, mitochondrial damage and apoptosis.2, 3 Accumulating evidence points to aberrant proteostasis and associated oxidative stress as key drivers of programmed cell death.3
The phosphorylation and dephosphorylation of the eukaryotic initiation factor, eIF2α, is a pivotal step in ‘integrated stress response’ (ISR)4 that is activated in response to many stresses, including DNA damage, ER stress, nutrient deprivation and oxidative stress. Four kinases, PERK, PKR, HRI and GCN2, responding to different stresses, catalyze the phosphorylation of serine-51 in eIF2α to repress global mRNA translation. However, phospho-eIF2α also facilitates the translation of selected mRNAs, such as ATF4 and CHOP, which orchestrate the transcription of numerous genes, including many required for protein synthesis,3 such as PPP1R15A, whose protein product, GADD34, assembles an eIF2α phosphatase that restores general protein synthesis and recovery from stress.5, 6
Sirtuin 1 (SIRT1)7, 8 catalyzes the deacetylation of both histones and non-histone substrates,9 including the nuclear proteins p53,10 nuclear factor-κB11 and Forkhead box protein O,12 and the non-nuclear proteins AKT113 and LKB1.14 By modulating their functions, SIRT1 has a key role in energy homeostasis and the cellular response to stress, particularly DNA damage, oxidative stress and aging.7 Thus, similar to ISR, the dysregulation of SIRT1 has been implicated in cancer, diabetes, heart disease and many aging-related diseases including Alzheimer’s disease.15, 16, 17, 18
Recent studies showed that ER stress induced SIRT1 expression,19 which in turn attenuated ER stress signaling and enhanced cell survival.20, 21, 22 Among the first stress proteins identified as a SIRT1 substrate was the X-box-binding protein 1 (XBP1s), the product of a mRNA spliced by IRE1 following ER stress.23 By deacetylating XBP1s, SIRT1 antagonized XBP1s-mediated transcription and reduced the cell’s tolerance for ER stress. Thus, SIRT1−/− MEFs displayed greater resistance to ER stress-mediated cell death than their WT counterparts. Other studies showed that ER stress reduced SIRT1 mRNA levels in brown adipocytes such that SIRT1 overexpression alleviated ER stress-induced cell death.24 The latter observation is consistent with studies by Ghosh et al.25, which noted SIRT1’s association with eIF2α (as well as PPP1R15A/GADD34 and PPP1R15B/CReP, components of cellular eIF2α phosphatases) that attenuated eIF2α phosphorylation. A later study in cardiomyocytes emphasized that SIRT1 principally targeted the ISR pathway and its association with eIF2α catalyzed its deacetylation at Ac-K141 and Ac-K143.26 Moreover, eIF2α acetylation paralleled its phosphorylation at Serine-51 and both modifications were enhanced by ER stress. Although the mechanistic basis for the collaboration between eIF2α acetylation and phosphorylation is still unclear, in these studies the loss of SIRT1 function aggravated ER stress-induced cardiac injury, whereas SIRT1 activation was cardioprotective. Additional study showed that SIRT1 activation by the endogenous active regulator of SIRT1 also attenuated eIF2α phosphorylation and regulated ribosome biogenesis.27 These data suggested that coordination of deacetylation and dephosphorylation of eIF2α may also be critical for ISR signaling and cytoprotection.
GADD34, which assembles a potent eIF2α phosphatase,6 interacted with SIRT1,25 but the functional consequence remains unclear. We identified several acetylated lysines in GADD34, raising the possibility that GADD34 deacetylation by SIRT1 might also regulate ISR. Conversely, SIRT1 is a phosphoprotein in cells28 with phosphorylations implicated in both positive and negative regulation of SIRT1. SIRT1 kinases include AMPK,29 Cyclin B/Cdk1,28 CK2,30 DYRK;31 JNK,32, 33, 34 CaMKKβ,35 HIPK2,36 Cdk5 (Bai et al.37) and mammalian target of rapamycin.38 However, SIRT1 phosphatases and the coordination of eIF2α deacetylation and dephosphorylation have not been investigated.
Our data showed that mutations that eliminated GADD34 acetylation at K315 and K322 did not have an impact on the assembly of eIF2α/GADD34/PP1α complex and the dephosphorylation of p-eIF2α. However, oxidative stress enhanced SIRT1 redistribution from the nucleus to the cytoplasm and promoted its binding to GADD34. The resulting complex, consisting of SIRT1, eIF2α, GADD34 and PP1α, synchronized the dephosphorylation of SIRT1-pSer47 and eIF2α-pSer51. The GADD34-mediated dephosphorylation enhanced SIRT1 deacetylase activity, measured both in vitro and in cells. Thus, our data suggested that the eIF2α/GADD34/PP1α/SIRT1 complex coordinates eIF2α deacetylation and dephosphorylation to control cell fate following oxidative stress.
Results
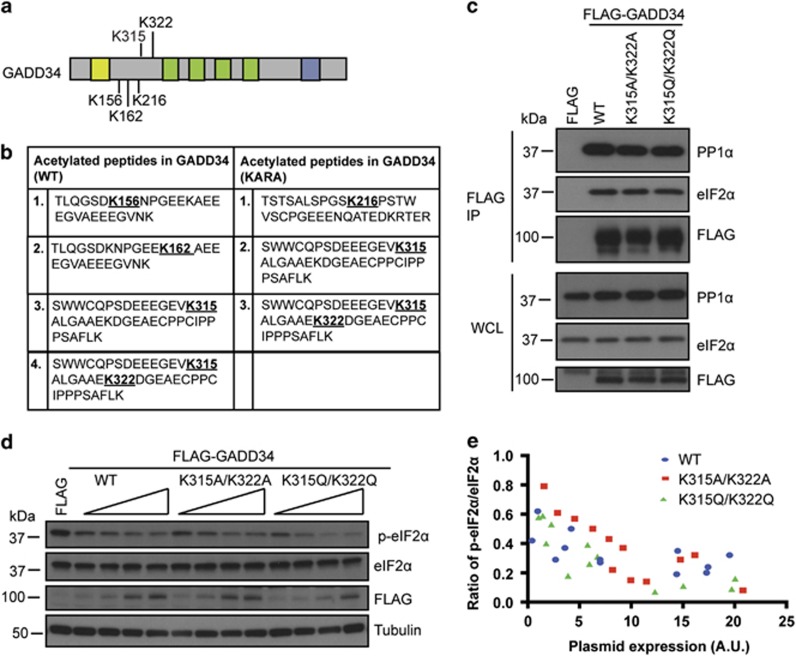
GADD34 is acetylated
Mass spectrometry of WT human FLAG-GADD34 and a mutant, KARA, which failed to bind PP1α,6 immunoprecipitated from HEK293 cells, identified multiple acetylated peptides. Data from independent experiments established GADD34 acetylation at five lysines (Figures 1a and b). However, the peptide containing Ac-K315 and/or Ac-K322 was most prominent in WT GADD34 after cells were treated with the SIRT1 inhibitor, nicotinamide. Interestingly, this peptide was also observed in KARA, independent of nicotinamide treatment. Substitution of alanines, K315A/K322A, or glutamines, K315Q/K322Q, did not alter GADD34’s ability to bind PP1α and eIF2α (Figure 1c), both being similarly recruited by WT and the mutant GADD34. Ectopic expression of WT, K315A/K322A or K315Q/K322Q displayed similar dose-dependent dephosphorylation of p-eIF2α (Figures 1d and e), suggested that GADD34 acetylation at K315 and K322, while potentially regulated by SIRT1, had no impact on assembly or function of the GADD34-containing eIF2α phosphatase.
Figure 1.
GADD34 is acetylated. (a) Schematic of the GADD34 protein with acetylated lysines (K). The N-terminal ER localization sequence is denoted by yellow box. The central PEST repeats are represented as green boxes and the C-terminal blue box represents the PP1-binding domain. (b) Mass spectrometry of FLAG-GADD34 (WT) or FLAG-GADD34 (KARA) immunoprecipitates (IP) from cells treated with or without 5 mM nicotinamide for 6 h identified acetylated peptides with sequences as shown. (c) HEK293 cells expressing FLAG-GADD34 (WT) and mutants, FLAG-K312A/K322A and FLAG-K315Q/K322Q, were subjected to immunoprecipitation using anti-FLAG antibody. Immunoprecipitates (IP) and whole cell lysates (WCL) were analyzed for endogenous eIF2α, PP1α and FLAG-GADD34 by immunoblotting. Molecular weight markers (kDa) are shown. (d) HEK293 cells expressing the FLAG epitope and increasing amounts of FLAG-GADD34 WT or FLAG-K315A/K322A and FLAG-K315Q/K322Q mutation were analyzed for eIF2α dephosphorylation. Cells were immunoblotting for eIF2α, p-eIF2α, FLAG-GADD34 and tubulin. (e) Quantitation of p-eIF2α/eIF2α ratio in cells expressing increasing amounts of the WT FLAG-GADD34, FLAG-K315A/K322A and FLAG-K315Q/K322Q (expressed in arbituary units, A.U.) from 4 independent experiments each consisting of 12 data points
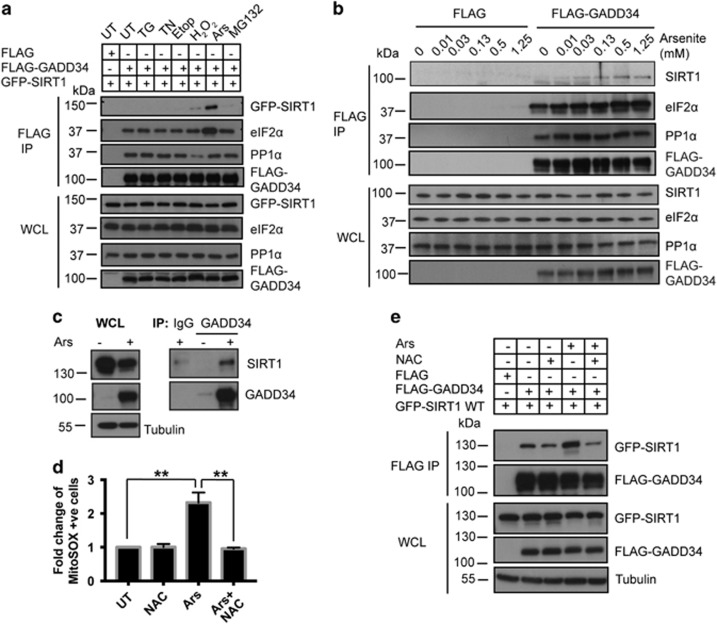
Arsenite promotes SIRT1 interaction with GADD34
Ghosh et al.25 showed that SIRT1 bound GADD34 but did not evaluate the impact of stresses that promote GADD34 expression. Thus, we coexpressed FLAG-GADD34 and GFP-SIRT1 in HEK293 cells and subjected these to different stresses (Figure 2a). Immunoprecipitation of FLAG-GADD34 from untreated (UT) cells confirmed the association of eIF2α and PP1α, known GADD34 interactors, but little or no SIRT1 was detected. Although ER stressors, tunicamycin (TN) and thapsigargin (TG) and DNA-damaging agent, etoposide (Etop) did not promote SIRT1 recruitment to FLAG-GADD34, arsenite (Ars) and to a lesser extent, H2O2, which induce an oxidative stress, and proteasome inhibition by MG132 resulted in detectable SIRT1 in the FLAG-GADD34 immunocomplexes. SIRT1 co-immunoprecipitation with FLAG-GADD34 showed a dose dependency with increasing Ars concentrations (Figure 2b). By contrast, GADD34 association with PP1α was unaltered by most stresses (Figure 2a) but some enhancement of eIF2α binding was seen with increasing Ars concentrations (Figures 2a and b). Finally, Ars increased endogenous GADD34 levels in WT MEFs while SIRT1 protein levels were consistently reduced. Regardless, endogenous SIRT1 was coimmunoprecipitated with endogenous GADD34 only from Ars-treated WT MEFs (Figure 2c). Ars targets thiol-mediated redox regulation and increased ROS, monitored using MitoSOX (Figure 2d). The increase in ROS production was abolished by the anti-oxidant, N-acetyl-L-cysteine (NAC). In parallel studies, Ars enhanced the binding of ectopically expressed GFP-SIRT1 to FLAG-GADD34, assessed using both anti-FLAG (Figure 2e) and anti-GFP (Supplementary Figure S1) antibodies. This binding was also reduced in Ars-treated cells by NAC. These data strongly supported the notion that Ars-induced oxidative stress not only enhanced GADD34 expression but also recruited SIRT1 to the GADD34 complex containing PP1α and eIF2α.
Figure 2.
Ars enhances SIRT1 association with GADD34. (a) HEK293 cells expressing FLAG-GADD34 were subjected to the following stresses for 1 h (hr): TG, TN, Etop, H2O2, Ars and MG132 before immunoprecipitations (IP) using anti-FLAG antibody. IP and WCL were immunoblotted for GFP-SIRT1, FLAG-GADD34, eIF2α and PP1α. Molecular weight markers (kDa) are shown. (b) HEK293 cells expressing the FLAG peptide or FLAG-GADD34 were subjected to increasing Ars concentrations for 1 h before IP using anti-FLAG. IP and WCL were immunoblotted for SIRT1, eIF2α, PP1α and FLAG-GADD34. (c) Anti-GADD34 immunoprecipitates from WT MEFs treated with Ars (50 μM, 5 h) or UT were immunoblotted for SIRT1 and GADD34. Control IgG antibody was included for control IP. Tubulin was used as loading control. (d) HEK293 cells, exposed to antioxidant, NAC (20 mM, 2 h), or Ars (0.5 mM, 1 h) or NAC (20 mM, 1 h) before Ars (0.5 mM, 1 h), were stained using mitochondrial ROS-detecting dye, MitoSOX. MitoSOX-stained cells were quantified by FACS and represented as fold change compared with UT HEK293 (mean±S.E.M., n=3). Sidak’s multiple comparisons test was used after two-way ANOVA to generate P-values (**P<0.01). (e) Anti-FLAG immunoprecipitations from HEK293 cells co-expressing FLAG-GADD34 and GFP-SIRT1 WT either UT (−) or following treatment with NAC, Ars or NAC and Ars as described in d were analyzed. IP and WCL were immunoblotted for GFP-SIRT1 and FLAG-GADD34. Tubulin was used as a loading control. Molecular weight markers (kDa) are shown
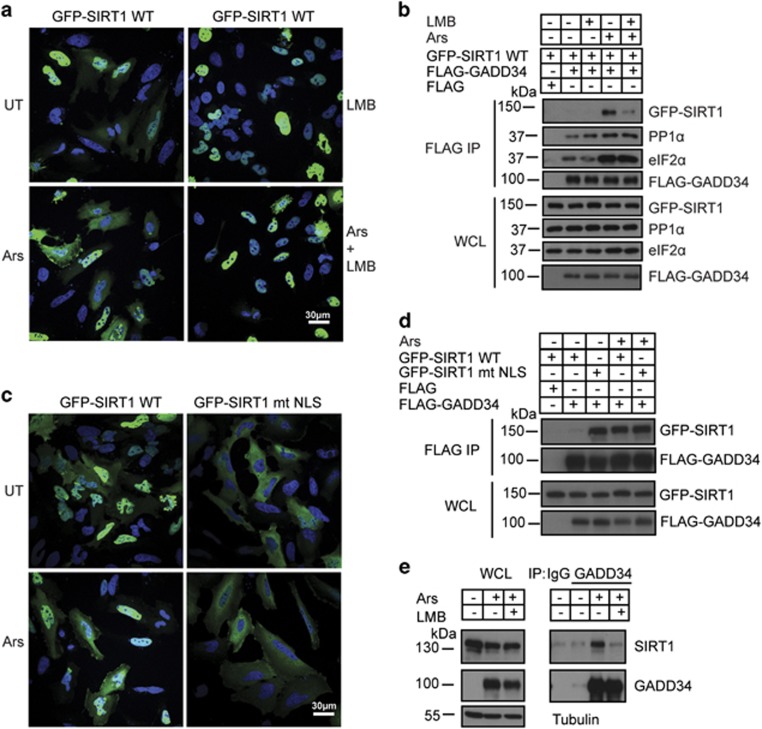
Cytosolic SIRT1 binds GADD34
GADD34 is primarily an ER-associated protein although a fraction of GADD34 also exists in cytoplasm.39 By contrast, SIRT1 is predominantly nuclear. This was confirmed by the subcellular distribution of GFP-SIRT1 expressed in UT HeLa cells (Figure 3a). However, oxidative stress was reported to enhance nucleocytoplasmic shuttling of SIRT1.40, 41 Indeed, Ars exposure enhanced the redistribution of GFP-SIRT1 to the cytoplasm in HeLa cells. Co-treatment with Leptomycin B (LMB), a nuclear export inhibitor, prevented the redistribution of GFP-SIRT1 to cytoplasm. When HEK293 cells expressing both GFP-SIRT1 (WT) and FLAG-GADD34 were exposed to Ars, GFP-SIRT1 binding to GADD34 was also attenuated by LMB (Figure 3b). However, LMB had no impact on PP1α or eIF2α binding to FLAG-GADD34. Following the mutation of the nuclear localization sequence (NLS) in SIRT1,40 GFP-SIRT1 mutant lacking the NLS (mt NLS) was largely cytoplasmic (Figure 3c). Remarkably, the association of GFP-SIRT1 mt NLS with FLAG-GADD34 occurred even in the absence of stress and Ars treatment has no impact on GFP-SIRT1 mt NLS binding to FLAG-GADD34 (Figure 3d). Immunoprecipitation using anti-GADD34 antibody also showed that Ars induced the association of endogenous SIRT1 with endogenous stress-induced GADD34 and this binding was also abolished by LMB (Figure 3e). Together, these results established that the cytoplasmic localization of SIRT1 elicited by Ars enabled GADD34 binding.
Figure 3.
Cytosolic SIRT1 Binds GADD34. (a) Subcelullar distribution of GFP-SIRT1 wild-type (WT) in HeLa cells is shown under UT conditions and after Ars (0.5 mM, 1 h) treatment. LMB (10 nM, 3 h), or LMB (10 nM, 2 h) pretreatment before Ars (0.5 mM, 1 h) exposure are also shown. GFP-SIRT1 is shown in green with nuclear staining by Hoescht 33342 in blue. Scale bar, 30 μm. (b) Anti-FLAG immunoprecipitations from HEK293 cells co-expressing FLAG-GADD34 and GFP-SIRT1, either UT (−) or following treatment with LMB, Ars or both LMB and Ars, as described in a, were analyzed for GFP-SIRT1, FLAG-GADD34, eIF2α and PP1α by immunoblotting IPs and WCL with appropriate antibodies. (c) Subcellular distribution of GFP-SIRT1 wild-type (WT) and mutant GFP-SIRT1 lacking the NLS (mt NLS) are shown in HeLa cells either UT or after Ars (0.5 mM, 1 h) treatment. GFP-SIRT1 is shown in green and nuclear staining by Hoescht 33342 in blue. Scale bar, 30 μm. (d) HEK293 cells coexpressing FLAG-GADD34 and either GFP-SIRT1 WT or GFP-SIRT1 mt NLS UT or following Ars treatment (0.5 mM, 1 h) were subjected immunoprecipitations using anti-FLAG. The GFP-SIRT1 proteins and FLAG-GADD34 in IP and WCL were analyzed by immunoblotting as described in methods. (e) Anti-GADD34 immunoprecipitates from WT MEFs either UT (−) or following treatment with Ars (50 μM, 5 h) or LMB (10 nM, 1 h) pretreatment before exposure to Ars (50 μM, 5 h) were immunoblotted with anti-SIRT1 and anti-GADD34 antibodies. Control IgG was included as control. Tubulin was used a loading control
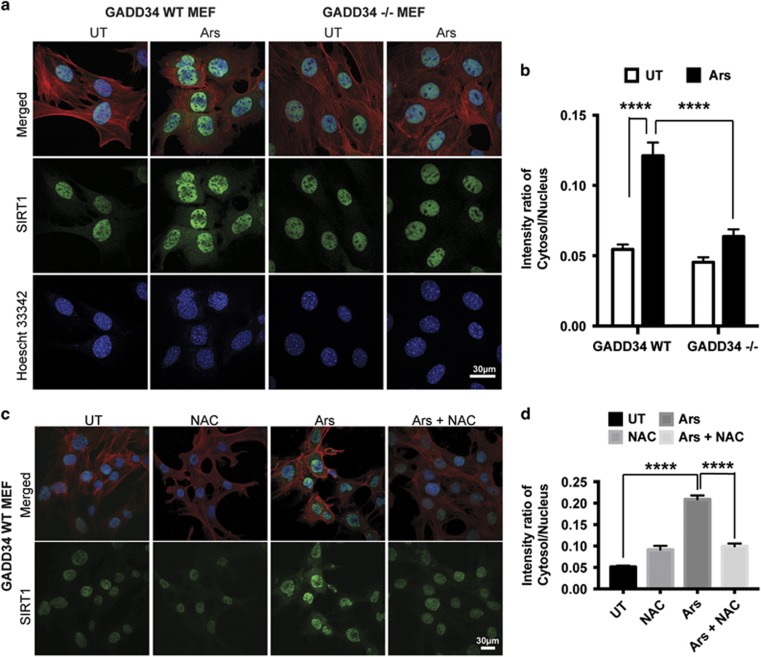
GADD34 contributes to cytoplasmic localization of SIRT1
Immunohistochemistry using anti-SIRT1 antibody established that endogenous SIRT1 was predominantly nuclear in WT MEFs but was partially redistributed to the cytoplasm following Ars treatment (Figure 4a). The ratio of cytoplasmic to nuclear SIRT1 was increased approximately 2.5-fold by Ars in WT MEFs (Figure 4b). Surprisingly, SIRT1 remained predominantly nuclear in the GADD34−/− MEFs in the absence or presence of Ars (Figures 4a and b). Finally, in WT MEFs, the Ars-mediated redistribution of SIRT1 to cytoplasm was prevented by NAC (Figures 4c and d). These data demonstrated that oxidative stress promoted the SIRT1 redistribution to cytoplasm and identified GADD34 as a key determinant of the cytoplasmic SIRT1 localization in MEFs.
Figure 4.
GADD34 contributes to SIRT1 localization in cytoplasm. (a) Immunohistochemistry assessed the subcellular distribution of SIRT1 (green) in WT and GADD34−/− MEFs either UT or following treatment with 50 μM Ars for 4 h. The nuclei were stained with Hoescht 33342 (blue) and cytoplasm outlined using Rhodamine-Phalloidin to stain filamentous actin (red). Scale bar, 30 μm. (b) The intensity of SIRT1 staining in nucleus and cytoplasm of WT and GADD34−/− MEFs before and after Ars treatment was quantified. (Mean±S.E.M., n≥15 cells). Tukey multiple comparison test was used after two-way ANOVA to generate P-values (****P<0.0001). (c) Immunohistochemistry assessed the subcellular distribution of SIRT1 (green) in WT MEFs either UT or NAC (20 mM, 5 h) or Ars (50 μM, 4 h), or pretreatment of NAC (20 mM, 1 h) before exposure to 50 μM Ars for 4 h. The nuclei were stained with Hoescht 33342 (blue) and cytoplasm outlined using Rhodamine-Phalloidin (red). Scale bar, 30 μm. (d) The intensity of SIRT1 staining in the nucleus and cytoplasm of WT MEFs subjected to various treatments as shown in c was quantified (mean±S.E.M., n≥70 cells). Tukey multiple comparison test was used after two-way ANOVA to generate P-values (****P<0.0001)
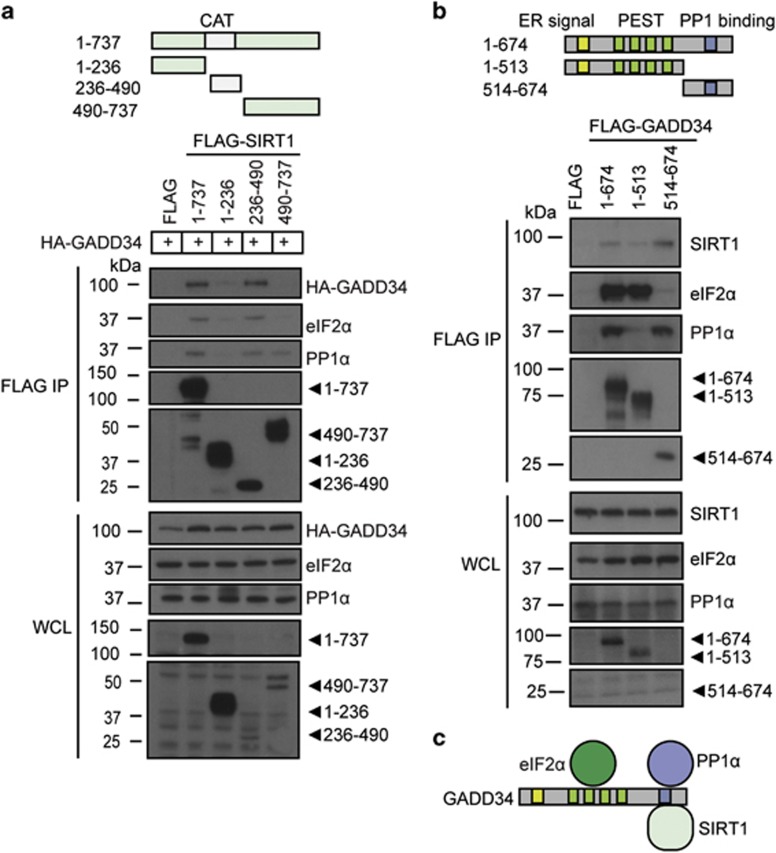
SIRT1 binds C-terminal PP1α-binding domain of GADD34
To define the interaction between SIRT1 and GADD34, fragments of FLAG-SIRT1 were coexpressed with HA-GADD34 in HEK293 cells. Following Ars exposure, immunoprecipitation with anti-FLAG antibody showed that HA-GADD34 bound full-length SIRT1 (1–737) and the catalytic domain, comprising of amino acids 236–490 (Figure 5a). No HA-GADD34 association was observed with the N- (1–236) and C-terminal (490–737) SIRT1 domains. Interestingly, the known GADD34 interactors,6 PP1α and eIF2α, were consistently observed in immunocomplexes containing GADD34 and SIRT1 (Figure 5a). In parallel experiments, co-immunoprecipitation of endogenous SIRT1 from Ars-treated HEK293 cells expressing FLAG-GADD34 fragments (Figure 5b) showed that SIRT1, bound the full-length and the C-terminal domain of GADD34 (513–674 amino acids) that also binds PP1α. 6 Choy et al.42 showed that eIF2α bound full-length GADD34 and the N-terminal fragment (1–513), containing PEST repeats, whereas others showed that the C-terminal PP1-binding domain of GADD34 also represented an eIF2α-binding site.43 These data highlighted GADD34’s ability to scaffold PP1α, eIF2α and SIRT1 (Figure 5c).
Figure 5.
Domains of GADD34 and SIRT1 that mediate their association. (a) Schematic of SIRT1 and its deletion mutants used to map the GADD34-binding site. HA-GADD34 was coexpressed with either empty FLAG vector, full-length FLAG-SIRT1 (1-737) and selected SIRT1 fragments in HEK293 cells, which were treated with Ars (0.5 mM, 1 h). Following immunoprecipitations using anti-FLAG, IP and WCL were immunoblotted for HA-GADD34, FLAG-SIRT1, eIF2α and PP1α. Molecular weight markers (kDa) are shown. (b) Schematic of GADD34 and its deletion mutants used to map the SIRT1-binding site. FLAG-GADD34 polypeptides were immunoprecipitated from HEK293 cells treated with 0.5 mM Ars for 1 h using anti-FLAG. IP and WCL were immunoblotted for SIRT1, eIF2α, PP1α and FLAG-GADD34. (c) Schematic summarizing the binding sites on GADD34 for SIRT1, eIF2α and PP1α is shown. The N-terminal ER localization sequence in GADD34 is shown as yellow box, the central PEST repeats as green boxes and the C-terminal PP1-binding domain as a blue box
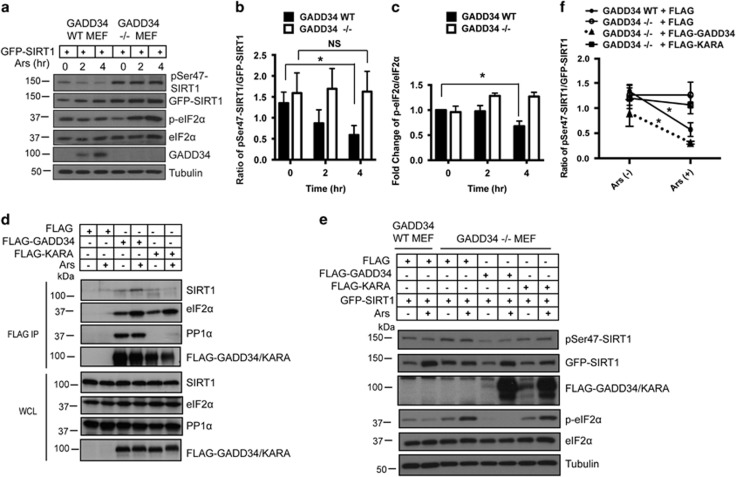
GADD34 facilitates dephosphorylation of SIRT1
To explore whether, similar to eIF2α, SIRT1 was a substrate of the GADD34/PP1α complex, we analyzed the phosphorylation of GFP-SIRT1, specifically Serine-47 phosphorylated by multiple kinases, in WT and GADD34−/− MEFs (Figure 6a). Following Ars exposure, p-Serine-47 levels decreased in a time-dependent manner in WT MEFs (Figure 6b) in parallel with GADD34 induction, which was also accompanied with p-eIF2α dephosphorylation (Figure 6c).5, 6 By comparison, phospho-serine 47 levels remained persistently high while p-eIF2α levels steadily increased in GADD34−/− MEFs in response to Ars (Figures 6a–c).
Figure 6.
GADD34 promotes dephosphorylation of phosphoserine-47 on SIRT1. (a) WT or GADD34−/− MEFs expressing GFP-SIRT1 were treated with 50 μM Ars for 0, 2 and 4 h before immunoblotting for phosphoserine-47 SIRT1 (pSer47-SIRT1), GFP-SIRT1, phosphoserine-51 eIF2α (p-eIF2α), eIF2α, GADD34 and tubulin. (b) Ratio of pSer47-SIRT1/GFP-SIRT1 level in WT or GADD34−/− MEFs treated with Ars for indicated times is shown as a bar graph (mean±S.E.M., n=3 independent experiments). Tuke’s multiple comparison test was used after two-way ANOVA to generate P-values (*P<0.05, NS denotes not significant). (c) Fold change of p-eIF2α/eIF2α in WT or GADD34−/− MEFs treated with Ars for indicated times is shown as a bar graph (mean±S.E.M., n=3 independent experiments). Dunnett’s multiple comparisons test was used after two-way ANOVA to generate P-values (*P<0.05, NS denotes not significant). (d) HEK293 cells expressing FLAG empty vector, FLAG-GADD34 or FLAG KARA were either UT (−) or Ars-treated (+)before immunoprecipitations using anti-FLAG conjugated beads. IP and WCLs were immunoblotted for SIRT1, eIF2α, PP1α and the FLAG-GADD34. (e) GFP-SIRT1 was coexpressed with either empty FLAG vector, FLAG-GADD34 or FLAG-KARA in GADD34−/− MEFs. As control, GFP-SIRT1 was co-expressed with empty FLAG vector in WT MEFs. Transfected MEFs were UT (−) or treated with 50 μM Ars for 4 h and immunoblotted as described in a. (f) Ratio of pSer47-SIRT1/GFP-SIRT1 in WT or GADD34−/− MEFs expressing various FLAG plasmids with or without Ars treatment (mean±S.E.M., n=3). Sidak multiple comparison test was used after two-way ANOVA to generate P-values (*P<0.05)
Although our experiments showed the binding of endogenous SIRT1, PP1α and eIF2α to FLAG-GADD34 in Ars-treated cells (Figure 2), to our surprise, KARA, a mutant GADD34 deficient in PP1α binding,6 showed low or no binding to SIRT1 in the presence or absence of Ars (Figure 6d). This provided a useful tool to assess the role of GADD34-bound PP1α in SIRT1 dephosphorylation. Thus, we expressed WT GADD34 and KARA along with GFP-SIRT1 in GADD34−/− MEFs and exposed them to Ars (Figure 6e). WT GADD34 but not KARA significantly reduced phospho-serine 47 levels in GFP-SIRT1 (Figures 6e and f). As anticipated, p-eIF2α levels were also reduced by WT GADD34 but not KARA in these cells (Figure 6e). One unexpected observation was that both WT GADD34 and KARA proteins were dramatically increased by Ars. While the molecular basis for upregulation of these ectopically expressed proteins is not known, our data suggest that post-transcriptional mechanisms activated by Ars accounted for this increase in GADD34 proteins. Regardless, our data established the requirement for GADD34 association with PP1α (and SIRT1) for dephosphorylation of phospho-serine 47.
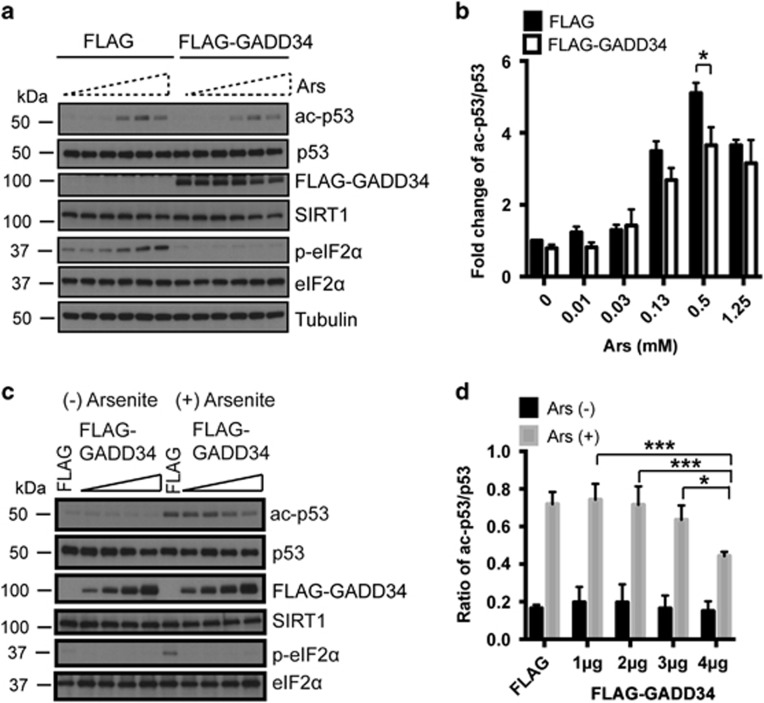
GADD34/PP1α increases SIRT1 deacetylase activity
Serine-47-phosphorylation inhibits SIRT1’s deacetylase function, potentially promoting the proteasomal degradation of SIRT1.34, 38 To investigate the changes in SIRT1 function elicited by GADD34/PP1α, we analyzed a known SIRT1 substrate, p53.10, 44 In control FLAG-expressing HEK293 cells, p53 was acetylated in a dose-dependent manner following Ars treatment (Figure 7a). Ectopic expression of WT FLAG-GADD34 significantly lowered the levels of acetylated-p53, particularly at higher Ars concentrations (Figures 7a and b). However, significant cell death was observed at 1.25 mM Ars. The complete abolition of p-eIF2α in cells expressing WT FLAG-GADD34 confirmed the function of GADD34/PP1α as an eIF2α phosphatase. Expression of increasing amounts of FLAG-GADD34 in the Ars-treated HEK293 cells also reduced acetylated p53 in a dose-dependent manner (Figures 7c and d). In other studies, we utilized siRNA targeting GADD34 to ‘knock down’ GADD34 levels in Ars-treated HEK293 cells (Supplementary Figure S2). This resulted in increases in p-eIF2α and acetylated p53, which were reversed by the co-expression of siRNA-resistant GADD34 plasmid. More direct evidence for changes in SIRT1 activity was obtained by assaying deacetylase activity in extracts from GADD34−/− MEFs expressing either WT FLAG-GADD34 or FLAG-KARA (Supplementary Figure S3). Immunoblotting for p-eIF2α confirmed that WT GADD34 but not KARA assembled an active eIF2α phosphatase (Supplementary Figure S3A). Similarly, SIRT1 deacetylase activity was increased in extracts from GADD34−/− MEFs expressing WT GADD34 but not KARA (Supplementary Figure S3B). Together, these data showed that dephosphorylation of phospho-serine 47 by GADD34/PP1α activated SIRT1.
Figure 7.
GADD34 enhances SIRT1’s deacetylase activity. (a) HEK293 cells expressing FLAG-GADD34 were exposed to increasing concentrations of Ars (1 h) and immunoblotted for acetylated p53 (ac-p53), p53, SIRT1, eIF2α, p-eIF2α, FLAG-GADD34 and Tubulin. (b) Quantification of the fold-change in ac-p53/p53 in HEK293 cells expressing FLAG or FLAG-GADD34 in the presence of increasing Ars concentrations (mean±S.E.M., n=3). Sidak’s multiple comparison test was used after two-way ANOVA to generate P-values (*P<0.05). (c) HEK293 cells expressing either FLAG or increasing amounts of the FLAG-GADD34 plasmid DNA were treated either without or with Ars (0.5 mM, 1 h) before immunoblotting for ac-p53, p53, SIRT1, eIF2α, p-eIF2α and FLAG-GADD34. (d) Ratio of ac-p53/p53 level in HEK293 cells expressing FLAG or increasing amounts of FLAG-GADD34 either UT (Ars−) or Ars-treated (+). (Mean±S.E.M., n=3). Tukey’s multiple comparison test was used after two-way ANOVA to generate P-values (*P<0.05 and ***P<0.001)
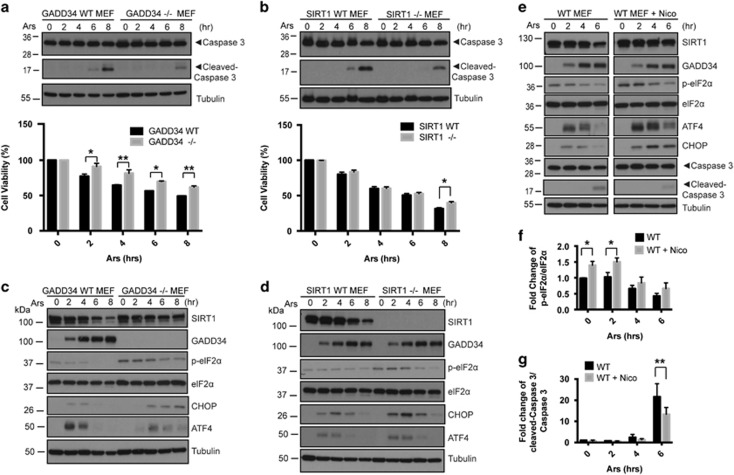
Loss of GADD34 or SIRT1 function elevates eIF2α phosphorylation and attenuates Ars-induced apoptosis
Prolonged Ars exposure resulted in apoptosis with the time-dependent activation of caspase-3 (Figure 8a, top panel) and reduced cell viability in WT MEFs (Figure 8a, bottom panel). Both outcomes were attenuated in the GADD34−/− MEFs (Figure 8a). The caspase-3 cleavage and reduced cell viability were similarly attenuated in SIRT1−/− MEFs (Figure 8b). SIRT1 protein levels were, however, identical in WT and GADD34−/− MEFs (Figure 8c). The loss of SIRT1 function also had no impact on GADD34 induction by Ars (Figure 8d). These data emphasized that loss of GADD34 did not alter SIRT1 expression and SIRT1 loss-of-function did not hinder the stress-mediated GADD34 induction. However, as previously reported in GADD34−/−45 and SIRT1−/− MEFs25 subjected to ER stress, persistent and elevated eIF2α phosphorylation together with protracted expression of downstream ISR genes, namely ATF4 and CHOP, were seen in both GADD34−/− and SIRT1−/− MEFs following Ars exposure (Figures 8c and d).
Figure 8.
Loss of function of GADD34 or SIRT1 attenuates apoptosis induced by Ars. (a) Top: WT and GADD34−/− MEFs were treated with 50 μM Ars for the indicated duration (hours) and immunoblotted for caspase 3, cleaved-caspase 3 and tubulin. Bottom: viability of WT and GADD34−/− MEFs was monitored over the time-course of the treatment with 50 μM Ars. (Mean±S.E.M., n=3). Sidak’s multiple comparison test was used after two-way ANOVA to generate P-values (*P<0.05, **P<0.01). Top: WT and SIRT1−/− MEFs were treated with 50 μM Ars for the indicated duration (hours) and cell lysates were immunoblotted for SIRT1, caspase 3, cleaved-caspase 3 and tubulin. (b) Bottom: viability of WT and SIRT1−/− MEFs was monitored over the time-course of the treatment with 50 μM Ars. (Mean±S.E.M., n=4 independent experiments). Sidak’s multiple comparison test was used after two-way ANOVA to generate P-values (*P<0.05). (c) WT and GADD34−/− MEFs were treated with 50 μM Ars for the indicated duration (hours) and immunoblotted for SIRT1, GADD34, p-eIF2α, eIF2α, ATF4, CHOP and tubulin. (d) WT and SIRT1−/− MEFs were treated with 50 μM Ars for the indicated duration (hours) and immunoblotted for SIRT1, GADD34, p-eIF2α, eIF2α, ATF4, CHOP and tubulin. (e) WT MEFs were treated with either 50 μM Ars for the indicated duration (hours) or pretreatment of Nicotinamide (Nico, 5 mM, overnight) following Ars exposure for the indicated duration (hours). Treated MEFs were immunoblotted with SIRT1, GADD34, p-eIF2α, eIF2α, ATF4, CHOP, caspase 3, cleaved-caspase 3 and tubulin. (f) Fold change of p-eIF2α/eIF2α in treated MEFs as described in e is shown as a bar graph (mean±S.E.M., n=3 independent experiments). Sidak’s multiple comparisons test was used after two-way ANOVA to generate P-values (*P<0.05). (g) Fold change of cleaved-caspase 3/caspase 3 in treated MEFs as described in e is shown as a bar graph (mean±S.E.M., n=3 independent experiments). Sidak’s multiple comparisons test was used after two-way ANOVA to generate P-values (**P<0.01)
To exclude adaptive changes resulting from loss of GADD34 or SIRT1 expression, we inhibited SIRT1 function using nicotinamide in WT MEFs (Figure 8e). As seen in SIRT1−/− MEFs, nicotinamide did not impair GADD34 induction by Ars. However, p-eIF2α levels were significantly elevated by nicotinamide (Figure 8f) and in turn resulted in the prolonged expression of downstream genes ATF4 and CHOP (Figure 6e). As also seen SIRT1−/− MEFs, nicotinamide significantly inhibited caspase-3 cleavage in the Ars-treated WT MEFs. These data showed that loss of function of SIRT1 gene or pharmacological inhibition of SIRT1 similarly elevated eIF2α phosphorylation and downstream ISR signaling with concomitant reduction in apoptosis following Ars exposure.
Discussion
Growing evidence supports a role for SIRT1 in the cellular response to stress. For example, oxidative stress reduced SIRT1 expression and suppressed SIRT1 activity12, 46, 47, 48 and reduction in SIRT1 function has been implicated in aging and aging-related diseases.49 Recent studies suggest that SIRT1 regulates ISR but the spectrum of stress response proteins regulated by SIRT1 are largely unknown. Current studies established that GADD34, a component of the stress-activated eIF2α phosphatase, shown to bind SIRT1,25 was acetylated on multiple lysines. Substituting two lysines, K315 and K322, whose acetylation was most consistently enhanced by SIRT1 inhibition, with residues that precluded acetylation, had no impact on assembly or function of the GADD34-containing eIF2α phosphatase. However, GADD34 protein is a short-lived protein and its stability controlled by various mechanisms, including shuttling on and off the ER membrane,39 phosphorylation50 and proteasomal degradation.51 Acetylation has been documented to regulate stability of many proteins52 and whether acetylation also controls to GADD34 protein stability will require further study.
SIRT1 is regulated by multiple mechanisms, including nucleocytoplasmic shuttling, phosphorylation at serines and threonines and protein turnover,41, 53, 54 all of which appear to be modulated by oxidative stress.46 Unlike an earlier report,25 our studies showed that oxidative stress, elicited by Ars (or H2O2) and countered by NAC, was an essential stimulus for SIRT1 binding to GADD34. Moreover, GADD34, an ER-localized and cytoplasmic protein,39 contributed to SIRT1’s cytoplasmic localization following Ars treatment as SIRT1 remained nuclear in Ars-treated GADD34−/− MEFs.40 Although inhibiting nuclear export with LMB attenuated SIRT1 binding, mutating the NLS sequence40 enhanced SIRT1 binding to GADD34 and precluded the requirement of Ars for interaction. These data strongly argued for the assembly of GADD34/SIRT1/PP1α/eIF2α complex in the cytoplasm.
Ars promoted GADD34 association with the SIRT1 catalytic domain. Conversely, SIRT1 bound to the C-terminal PP1-binding domain in GADD34 that also scaffolded PP1α, suggesting potential substrate-enzyme relationships. Our data hinted at possible cooperativity between GADD34 and PP1α in recruiting SIRT1 as the mutant GADD34 (KARA) showed severely reduced SIRT1 association, although direct association between SIRT1 and PP1α could not be demonstrated. By contrast, Ars not only enhanced GADD34 binding to SIRT1 but also its association with eIF2α, which independently bound SIRT1 (Ghosh et al.25) and GADD34.6, 42
The function of the SIRT1/GADD34/PP1α/eIF2α complex was most likely to coordinate the dephosphorylation of SIRT1 (phosphoserine-47) and eIF2α (phosphoserine-51) as Ars-induced GADD34 expression resulted in the parallel dephosphorylation of p-eIF2α (phosphoserine-51) and SIRT1 (phosphoserine-47). Although both events were absent in GADD34−/− MEFs, they could be restored by reexpression of WT GADD34 but not the phosphatase-deficient mutant, KARA, supporting a role for GADD34/PP1α as both an eIF2α and SIRT1 phosphatase. Finally, GADD34/PP1α-mediated dephosphorylation of SIRT1 enhanced its deacetylase activity, measured in vitro and in cells. Ars is among the most potent inducers of endogenous GADD34.51 The dramatic elevation of ectopically expressed GADD34 proteins by Ars (Figure 6e) may also point to oxidative stress-mediated proteasome inhibition55, 56 as a key factor in driving up high levels of GADD34 in cells experiencing oxidative stress,51 with the potential to prematurely terminate ISR signaling. Thus, we propose a model for the role of GADD34, which scaffolds of PP1α and SIRT1 in proximity to eIF2α, in controlling the duration of eIF2α phosphorylation/dephosphorylation and ISR signaling. While prior studies established that eIF2α acetylation parallels its phosphorylation at serine-51,26 the current work demonstrates the dephosphorylation and activation of SIRT1 by GADD34/PP1α complex, highlighting a necessary and potentially rate-limiting step that coordinates eIF2α deacetylation with its dephosphorylation, delaying the termination of ISR and ensuring effective downstream signaling (Figure 9). Consistent with this model, the loss of function of either SIRT1 or GADD34 elevated eIF2α phosphorylation and prolonged downstream signaling. Critical support for this model comes from the analysis of SIRT1−/− MEFs, where despite effective GADD34 induction by Ars, elevated and persistent eIF2α phosphorylation was seen. As GADD34 remained competent to assemble the GADD34/PP1α/eIF2α complex in SIRT1−/− MEFs, these experiments argued for an essential role for SIRT1-mediated eIF2α deacetylation in facilitating the subsequent dephosphorylation of phospho-Serine 51 by the GADD34-associated eIF2α phosphatase.
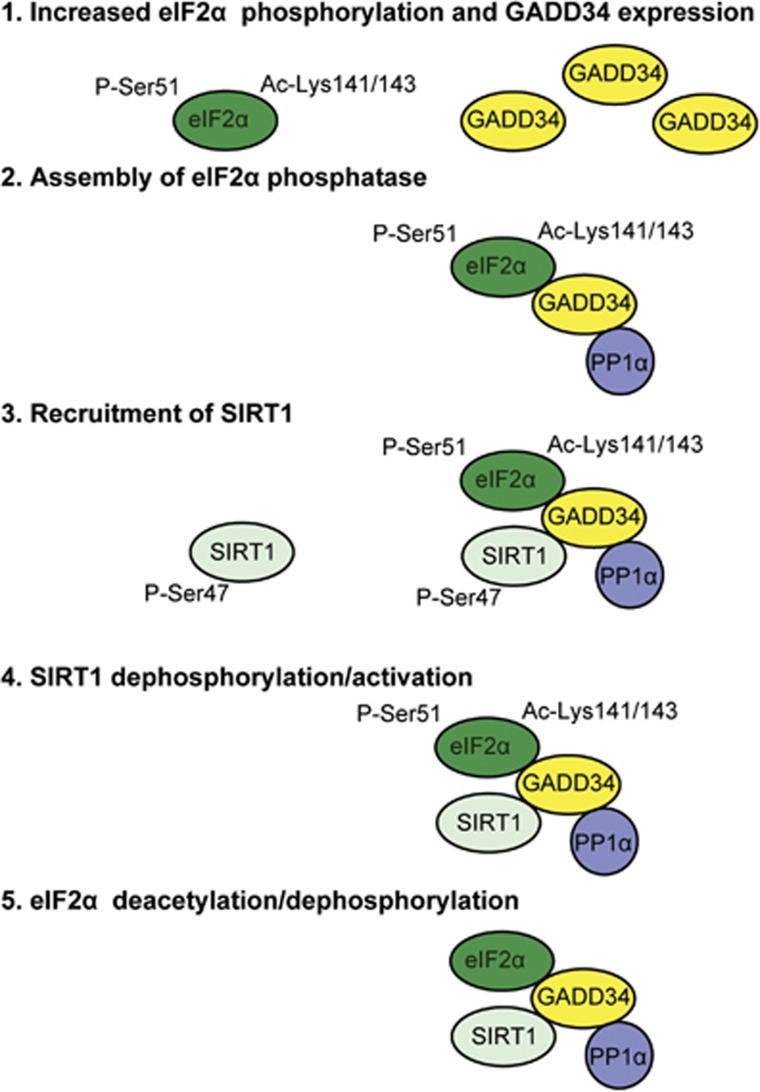
Figure 9.
Oxidative stress regulates eIF2α phosphorylation and dephosphorylation (The 5-STEP Model). Oxidative stress promotes eIF2α phosphorylation at serine-51, which is enhanced by the acetylation of lysines-141/143.26 In addition to increasing p-eIF2α, oxidative stress engages other as yet unknown mechanisms to elevate cellular GADD34 to much higher levels than seen with other stresses (STEP 1). Assembly of the GADD34/p-eIF2α/PP1α complex (STEP 2) enables eIF2α dephosphorylation and terminates ISR signaling. To ensure adequate downstream ISR signaling, we propose that acetylation of eIF2α at lysines 141/143 impedes the dephosphorylation by GADD34-bound PP1α. However, oxidative stress also recruits p-SIRT1, whose phosphorylation at serine-47 is enhanced by stress (STEP 3). Assembly of the new SIRT1/GADD34/eIF2α/PP1α complex enables SIRT1 dephosphorylation by the GADD34-bound PP1α and the resulting SIRT1 activation (STEP 4) catalyzes eIF2α deacetylation. This removes the brake on GADD34-bound PP1α and accelerates eIF2α dephosphorylation (STEP 5) to terminate ISR signaling. Thus, the coordination of eIF2α deacetylation with its dephosphorylation generates a delay that ensures the activation of the complex transcriptional and translational program known as ISR that is critical for cell survival
Although SIRT1 was shown to catalyze eIF2α deacetylation on K141 and K143,26 we were unable to monitor changes in eIF2α (or GADD34) acetylation, using commercially available anti-acetylated-lysine antibodies. Thus, other approaches were utilized in current studies to establish the cooperation of SIRT1 and PP1α in regulating eIF2α dephosphorylation. It should also be noted that the contributions of acetylations of GADD34 and/or other components of the eIF2α phosphatase complex in modulating eIF2α dephosphorylation cannot be excluded. It is noteworthy that PP1α is acetylated on multiple lysines (www.phosphosite.org). We previously reported that PP1α binds selected histone deacetylases (HDACs), an association that was disrupted by HDAC inhibitors,57 whereas we have been unable to conclusively show a direct association between PP1α and SIRT1. This likely points to HDACs rather than SIRT1 as potential PP1α deacetylase(s).
Finally, the loss of function of either SIRT1 or GADD34 attenuated apoptosis triggered by Ars. Future studies identifying the spectrum of changes in transcriptome and translatome in GADD34−/− and SIRT1−/− cells following oxidative stress or appropriate disease models should highlight the common pathways for cell fate determination and help to strengthen the physiological relevance of crosstalk between GADD34 and SIRT1 in the cellular stress response and disease.
Materials and methods
cDNAs
Plasmid pXJ40 containing the human GADD34 cDNA with N-terminal FLAG epitope was used for the expression of GADD34. The cDNAs for GADD34 and its deletion mutants (1–513 and 514–674) were obtained from pSG5-FLAG-GADD34.6 pSG5-FLAG-GADD34 and pSG5-FLAG-KARA were previously described.6 Point mutations in FLAG-GADD34 (1-674) (K315A/K322A; K315Q/K322Q) were also introduced using site-directed mutagenesis method following Agilent protocol. pEGFP-N3-SIRT1 and the mt NLS were kind provided by Yoshiyuki Horio (Sapporo Medical University, Sapporo, Japan).40 Deletion mutant of SIRT1 (1–236, 236–490 and 490–737) were subcloned from pEGFP-N3-SIRT1.
Antibodies
The following antibodies were used in this study. FLAG (Sigma Aldrich, St. Louis, MO, USA: F7425; 1:1000), GFP (Santa Cruz, Dallas, TX, USA: sc-8334; 1:1000), eIF2α (Santa Cruz: sc-11386; 1:1000), phospho-eIF2α (Ser51) (Invitrogen, Waltham, MA, USA, 44-728G; 1:1000), PP1α (Santa Cruz: sc-6104; 1:1000), mouse GADD34 (Santa Cruz, sc-825; 1:1000), mouse and human GADD34 (Proteintech, Rosemont, IL, USA, 10449-1-AP; 1:1000) human SIRT1 (Santa Cruz, sc-15404; 1:1000), mouse SIRT1 (Millipore, Temecula, CA, USA, 07-131; 1:2000), phosphoserine-47 SIRT1 (Cell Signaling, Beverly, MA, USA, 2314; 1:1000), acetylated-p53 (K379) (Cell Signaling, 2570; 1:1000), p53 (Santa Cruz, DO-1, sc106; 1:1000), caspase 3 and cleaved caspase 3 (Cell Signaling, 9662; 1:1000), ATF4 (Cell Signaling, 11815; 1:1000), CHOP (Cell Signaling, 5554; 1:1000) and tubulin (T5168; Sigma, St. Louis, MO, USA; 1:10 000).
Cell culture, transfection and drug treatment
HEK293 cells were grown in RPMI supplemented with 4500 mg/l glucose, 10% fetal bovine serum (Hyclone, Gibco, Thermo Scientific, Waltham, MA, USA) and 2 mM L-Glutamine (Gibco, Life Technologies, Waltham, MA, USA). HeLa cells were grown in minimum essential media (MEM) (Gibco, Life Technologies) supplemented with 4500 mg/l glucose, 10% fetal bovine serum (Hyclone/GE Healthcare, Pittsburg, PA, USA), 2 mM L-glutamine (Gibco/Life Technologies), 1 mM sodium pyruvate (Gibco/Life Technologies), 1 × MEM Non-Essential Amino Acids (Gibco/life Technologies). GADD34+/+ and GADD34−/− cells were obtained through serial passage of SV40 T-antigen transfected MEFs derived from WT and GADD34−/− mice.45 MEFs derived from SIRT1+/+ and −/− mice were kindly provided by Zhao Yingming (University of Chicago).58 MEFs were maintained in Dulbecco’s modified Eagle medium (Gibco, Life Technologies) supplemented with 4500 mg/l glucose, 10% fetal bovine serum (Hyclone/GE Healthcare), 100 U/ml penicillin/streptomycin (Gibco/life Technologies), 1 × MEM non-essential amino acids (Gibco/life Technologies) and 55 μM 2-Mercaptoethanol (Sigma). All cells were grown at 37 °C in an incubator containing 5% (v/v) CO2 atmosphere.
Transfections of HEK293 cells and HeLa cells with plasmid DNAs were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
MEFs (1.5 × 106 cells in 100 μl) were subjected to electroporation with 5 μg plasmid DNA using Neon Transfection System (Invitrogen). Transiently transfected HEK293 cells were treated with 200 nM Okadaic Acid (Enzo Lifesciences, Tokyo, Japan, ALX-350-003-C100) for 1 h before cell lysis and immunoblotting for p-eIF2α. Transiently transfected HEK293 cells were treated for 1 h with either 5 μM TG (Sigma/DMSO), 10 μg/ml TN (Calbiochem, Temecula, CA, USA/DMSO), 20 μM Etop (Sigma/water), 0.5 mM Ars (Sodium Ars/water) or 5 μM MG132 (Sigma/DMSO), unless otherwise stated before cell lysis followed by immunoblotting or immunoprecipitations. Transiently transfected HEK293 cells were treated with 10 nM LMB (Sigma) for 3 h or pre-treated LMB for 2 h before exposure with Ars for 1 h before cell lysis and immunoprecipitation.
Transiently transfected HEK293 cells were treated with 0.5 mM Ars for 1 h, 20 mM NAC (Sigma) for 2 h or pretreated with NAC for 1 h before exposure with Ars for 1 h before cell lysis and immunoprecipitation.
Electroporated MEFs were treated with 50 μM Ars and collected at the stated time points, while Ars stress profile in the various genotype MEFs was monitored in cells treated with 50 μM Ars and harvested at the indicated time points. Ars stress profile of WT MEFs with inhibited SIRT1 activity was analyzed by treating MEFs with 5 mM Nicotinamide (Sigma), overnight, before co-treatment with Ars at the different time point.
Immunoblotting
Cells were washed with phosphate-buffered saline (PBS) and collected in lysis buffer (25 mM HEPES pH7.5, 150 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 5% (v/v) Glycerol, 1 mM Na3VO4, 0.5% (w/v) TritonX100, PhoSTOP (Roche Applied Science, Indianapolis, IN, USA) and Protease inhibitors (Roche Applied Science). Crude lysates were precleared by centrifugation (10 000 × g for 15 min at 4 °C) and protein content determined using Bradford assay (Biorad, Hercules, CA, USA). Lysates were boiled in sample buffer containing 37.5 mM Tris-HCl pH 6.8, 1.5% (v/v) β-mercaptoethanol, 0.075% (v/v) bromophenol blue and 37.5% (v/v) glycerol for 5 min. Equal amounts of protein were subjected to SDS-PAGE followed by immunoblotting.
Immunoprecipitation
All immunoprecipitations were performed as previously described.59 In brief, cells were lysed in cell lysis buffer (25 mM HEPES pH7.5, 150 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 5% (v/v) Glycerol, 1 mM Na3VO4, 0.5% (w/v) Triton X-100, PhoSTOP (Roche Applied Science) and Protease inhibitors (Roche Applied Science), and precleared by centrifugation (10 000 × g for 20 min at 4 °C) before incubation with either mouse M2 anti-FLAG beads (Sigma-Aldrich), GFP-affinity beads (Chromotek, Germany) or anti-GADD34 (sc-825) antibody in the presence of hydrated protein G-conjugated agarose beads overnight at 4 °C. After extensive washing with lysis buffer devoid of protease inhibitors, immunoprecipitates were boiled in sample buffer and subjected to SDS-PAGE and immunoblotting.
Immunohistochemistry
GFP-SIRT1-expressing HeLa cells were treated with 50 μM Ars for 4 h before fixation in 4% (v/v) paraformaldehyde for 20 min at room temperature. Fixed cells were stained with Hoechst 33342 to highlight the nucleus and Rhodamine-conjugated Phalloidin (ThermoFisher Scientific, Waltham, MA, USA) to show filamentous actin (F-actin). Drug-treated MEFs were incubated with anti-SIRT1 antibody (Millipore, 07-131; 1:200) overnight at 4 °C followed by anti-rabbit secondary antibodies conjugated with either Alexafluor-488 or -546 (Invitrogen) for 2 h at room temperature. Rhodamine-conjugated Phalloidin was used to visualize F-actin. Images were acquired using a CoolSNAP HQ2 camera (CCD) adapted to Olympus IX71 Inverted Epi-Fluorescent Microscope. Fluorescence intensities for cytosolic and nuclear SIRT1 were quantified in Image J software.
Measurement of ROS level by flow cytometry
Mitochondrial ROS level in HEK293 cells subjected to different drug treatment was assessed by MitoSOX (Thermo Fisher Scientific). Approximately 1 × 106 cells were incubated with 5 μM MitoSOX for 30 min at 37 °C and washed away with PBS+2% FBS. Cells were then stained with viable dye, 7-Amino-Actinomycin D (7-AAD) (Beckman Coulter, Brea, CA, USA) for 20 min at 4 °C and cells were washed twice with PBS+2% FBS, resuspended in 300 μl PBS+2% FBS and analyzed by flow cytometry (BD LSRFortessa, BD Biosciences, Franklin Lakes, NJ, USA). Excitation for MitoSOX was set at 510 nm and fluorescence detection at 580 nm, whereas excitation for 7-AAD was set at 480 nm and fluorescence detection at 655 nm. Percentage of viable (7-AAD positive) and MitoSOX-stained cells was determined in each treatment group.
Mass spectrometry
Mass spectrometry of immunoprecipitated WT FLAG-GADD34 and KARA, a mutant GADD34 unable to bind PP1, expressed in HEK293 cells was undertaken as previously described.50 Cells expression either WT FLAG-GADD34 or FLAG-KARA treated with or without with 5 mM nicotinamide, a SIRT1 inhibitor for 6 h and analyzed for peptides containing acetylated lysines.
CellTiter-Glo viability assay
Cells plated in 96-well plates were treated with 50 μM Ars for the stated times and lysed with CellTiter-Glo luminescent cell viability assay reagent (Promega) according to manufacturer’s instructions. Luminescence was read using a Tecan Infinite M200 microplate reader. The percent viability of cells following Ars treatment was calculated relative to UT cells.
Acknowledgments
We thank Dr Sudipto Bari for his technical assistance in flow cytometry and Dr Meng Choy for technical contribution. We also thank Yoshiyuki Horio (Sapporo Medical University) for providing DNA plasmids, pEGFP-N3-SIRT1 and the mt NLS. We also thank Zhao Yingming (University of Chicago) for providing SIRT1+/+ and −/− MEFs. This work was funded by A Translational Clinical Research Flagship Award entitled ‘National Parkinson’s Disease Translational Clinical Research Programme’ from National Medical Research Council of Singapore (to SS), Duke-NUS start-up funding provided by the Singapore Ministry of Health (to SS), and two A*STAR/BMRC TCRP (Translational and Clinical Research Partnership) Grants (to SS). CWG was supported by a Duke-NUS graduate fellowship.
Author contributions
ICL had a key role in the design of the study, undertook majority of the experiments with help from XYH and participated extensively in writing the manuscript. Additional experimental help was provided by SEG, CWG, JRS, KKLP, WL and PY. NSKS provided critical oversight and interpretation for all mass spectrometry experiments. SS participated in the study design, data interpretation and had a major role in writing the manuscript.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by N Chandel
The authors declare no conflict of interest.
Supplementary Material
References
- Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 2016; 529: 326–335. [DOI] [PubMed] [Google Scholar]
- Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R et al. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev 2004; 18: 3066–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol 2013; 15: 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep 2016; 17: 1374–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol 2001; 153: 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush MH, Weiser DC, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 to the endoplasmic reticulum and promotes dephosphorylation of the subunit of eukaryotic translation initiation factor 2. Mol Cell Biol 2003; 23: 1292–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol 2010; 5: 253–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem 2004; 73: 417–435. [DOI] [PubMed] [Google Scholar]
- Martinez-Redondo P, Vaquero A. The diversity of histone versus nonhistone sirtuin substrates. Genes Cancer 2013; 4: 148–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 2001; 107: 137–148. [DOI] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J 2004; 23: 2369–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 2004; 303: 2011–2015. [DOI] [PubMed] [Google Scholar]
- Sundaresan NR, Pillai VB, Wolfgeher D, Samant S, Vasudevan P, Parekh V et al. The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Sci Signal 2011; 4: ra46. [DOI] [PubMed] [Google Scholar]
- Lan F, Cacicedo JM, Ruderman N, Ido Y. SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP-activated protein kinase activation. J Biol Chem 2008; 283: 27628–27635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Fang D. The roles of SIRT1 in cancer. Genes Cancer 2013; 4: 97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima S, Sadoshima J. The role of sirtuins in cardiac disease. Am J Physiol Heart Circ Physiol 2015; 309: H1375–H1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Jiang X, Ma H, Wang Y, Xue P, Liu Y. SIRT1 and insulin resistance. J Diabetes Complications 2016; 30: 178–183. [DOI] [PubMed] [Google Scholar]
- Massudi H, Grant R, Braidy N, Guest J, Farnsworth B, Guillemin GJ. Age-associated changes in oxidative stress and NAD+ metabolism in human tissue. PLoS ONE 2012; 7: e42357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga T, Suico MA, Shimasaki S, Watanabe E, Kai Y, Koyama K et al. Endoplasmic reticulum (ER) stress induces Sirtuin 1 (SIRT1) expression via the PI3K-Akt-GSK3beta signaling pathway and promotes hepatocellular injury. J Biol Chem 2015; 290: 30366–30374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xu S, Giles A, Nakamura K, Lee JW, Hou X et al. Hepatic overexpression of SIRT1 in mice attenuates endoplasmic reticulum stress and insulin resistance in the liver. FASEB J 2011; 25: 1664–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev Cell 2005; 9: 605–615. [DOI] [PubMed] [Google Scholar]
- Jung TW, Lee KT, Lee MW, Ka KH. SIRT1 attenuates palmitate-induced endoplasmic reticulum stress and insulin resistance in HepG2 cells via induction of oxygen-regulated protein 150. Biochem Biophys Res Commun 2012; 422: 229–232. [DOI] [PubMed] [Google Scholar]
- Wang FM, Chen YJ, Ouyang HJ. Regulation of unfolded protein response modulator XBP1s by acetylation and deacetylation. Biochem J 2011; 433: 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Gu H, Gan L, Xu Y, Feng F, Saeed M et al. Reducing Smad3/ATF4 was essential for Sirt1 inhibiting ER stress-induced apoptosis in mice brown adipose tissue. Oncotarget 2017; 8: 9267–9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh HS, Reizis B, Robbins PD. SIRT1 associates with eIF2-alpha and regulates the cellular stress response. Sci Rep 2011; 1: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prola A, Silva JP, Guilbert A, Lecru L, Piquereau J, Ribeiro M et al. SIRT1 protects the heart from ER stress-induced cell death through eIF2alpha deacetylation. Cell Death Differ 2017; 24: 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JR, Willis AE, Milner J. Active regulator of SIRT1 is required for ribosome biogenesis and function. Nucleic Acids Res 2013; 41: 4185–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Maier B, Koclega KD, Chruszcz M, Gluba W, Stukenberg PT et al. Phosphorylation regulates SIRT1 function. PLoS ONE 2008; 3: e4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AW, Liu P, Inuzuka H, Gao D. SIRT1 phosphorylation by AMP-activated protein kinase regulates p53 acetylation. Am J Cancer Res 2014; 4: 245–255. [PMC free article] [PubMed] [Google Scholar]
- Zschoernig B, Mahlknecht U. Carboxy-terminal phosphorylation of SIRT1 by protein kinase CK2. Biochem Biophys Res Commun 2009; 381: 372–377. [DOI] [PubMed] [Google Scholar]
- Guo X, Williams JG, Schug TT, Li X. DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J Biol Chem 2010; 285: 13223–13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford J, Ahmed S, Allison S, Jiang M, Milner J. JNK2-dependent regulation of SIRT1 protein stability. Cell Cycle 2008; 7: 3091–3097. [DOI] [PubMed] [Google Scholar]
- Nasrin N, Kaushik VK, Fortier E, Wall D, Pearson KJ, de Cabo R et al. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS ONE 2009; 4: e8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Kheterpal I, Kennedy N, Davis RJ, Ye J. Sirtuin 1 (SIRT1) protein degradation in response to persistent c-Jun N-terminal kinase 1 (JNK1) activation contributes to hepatic steatosis in obesity. J Biol Chem 2011; 286: 22227–22234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Chen Z, Zhang F, Cui X, Sun W, Geary GG et al. Ca2+/calmodulin-dependent protein kinase kinase beta phosphorylation of Sirtuin 1 in endothelium is atheroprotective. Proc Natl Acad Sci USA 2013; 110: E2420–E2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad E, Polonio-Vallon T, Meister M, Matt S, Bitomsky N, Herbel C et al. HIPK2 restricts SIRT1 activity upon severe DNA damage by a phosphorylation-controlled mechanism. Cell Death Differ 2016; 23: 110–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai B, Liang Y, Xu C, Lee MY, Xu A, Wu D et al. Cyclin-dependent kinase 5-mediated hyperphosphorylation of sirtuin-1 contributes to the development of endothelial senescence and atherosclerosis. Circulation 2012; 126: 729–740. [DOI] [PubMed] [Google Scholar]
- Back JH, Rezvani HR, Zhu Y, Guyonnet-Duperat V, Athar M, Ratner D et al. Cancer cell survival following DNA damage-mediated premature senescence is regulated by mammalian target of rapamycin (mTOR)-dependent Inhibition of sirtuin 1. J Biol Chem 2011; 286: 19100–19108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Brush MH, Choy MS, Shenolikar S. Association with endoplasmic reticulum promotes proteasomal degradation of GADD34 protein. J Biol Chem 2011; 286: 21687–21696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem 2007; 282: 6823–6832. [DOI] [PubMed] [Google Scholar]
- Hwang JW, Yao H, Caito S, Sundar IK, Rahman I. Redox regulation of SIRT1 in inflammation and cellular senescence. Free Radic Biol Med 2013; 61: 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy MS, Yusoff P, Lee IC, Newton JC, Goh CW, Page R et al. Structural and functional analysis of the GADD34:PP1 eIF2alpha phosphatase. Cell Rep 2015; 11: 1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas M, Vasconcelos G, Dever TE. An eIF2alpha-binding motif in protein phosphatase 1 subunit GADD34 and its viral orthologs is required to promote dephosphorylation of eIF2alpha. Proc Natl Acad Sci USA 2015; 112: E3466–E3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 2001; 107: 149–159. [DOI] [PubMed] [Google Scholar]
- Reid DW, Tay AS, Sundaram JR, Lee IC, Chen Q, George SE et al. Complementary roles of GADD34- and CReP-containing eukaryotic initiation factor 2alpha phosphatases during the unfolded protein response. Mol Cell Biol 2016; 36: 1868–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Kwon YE, Song JO, Bae SJ, Seol JH. CHFR negatively regulates SIRT1 activity upon oxidative stress. Sci Rep 2016; 6: 37578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caito S, Rajendrasozhan S, Cook S, Chung S, Yao H, Friedman AE et al. SIRT1 is a redox-sensitive deacetylase that is post-translationally modified by oxidants and carbonyl stress. FASEB J 2010; 24: 3145–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra M, Santini MP, Martinez C, Pazienza V, Claycomb WC, Giuliani A et al. mIGF-1/JNK1/SirT1 signaling confers protection against oxidative stress in the heart. Aging Cell 2012; 11: 139–149. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Yankner BA. The aging stress response. Mol Cell 2010; 40: 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Jeyaraman K, Yusoff P, Shenolikar S. Phosphorylation at tyrosine 262 promotes GADD34 protein turnover. J Biol Chem 2013; 288: 33146–33155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush MH, Shenolikar S. Control of cellular GADD34 levels by the 26S proteasome. Mol Cell Biol 2008; 28: 6989–7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron C, Boyault C, Khochbin S. Regulatory cross-talk between lysine acetylation and ubiquitination: role in the control of protein stability. Bioessays 2005; 27: 408–415. [DOI] [PubMed] [Google Scholar]
- Buler M, Andersson U, Hakkola J. Who watches the watchmen? Regulation of the expression and activity of sirtuins. FASEB J 2016; 30: 3942–3960. [DOI] [PubMed] [Google Scholar]
- Sun L, Fang J. Macromolecular crowding effect is critical for maintaining SIRT1's nuclear localization in cancer cells. Cell Cycle 2016; 15: 2647–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yen J, Kaiser P, Huang L. Regulation of the 26S proteasome complex during oxidative stress. Sci Signal 2010; 3: ra88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiken CT, Kaake RM, Wang X, Huang L. Oxidative stress-mediated regulation of proteasome complexes. Mol Cell Proteomics 2011; 10 R110 006924. doi:10.1074 mcp.R110.006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush MH, Guardiola A, Connor JH, Yao TP, Shenolikar S. Deactylase inhibitors disrupt cellular complexes containing protein phosphatases and deacetylases. J Biol Chem 2004; 279: 7685–7691. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhao W, Yang JS, Cheng Z, Luo H, Lu Z et al. Quantitative acetylome analysis reveals the roles of SIRT1 in regulating diverse substrates and cellular pathways. Mol Cell Proteomics 2012; 11: 1048–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IC, Leung T, Tan I. Adaptor protein LRAP25 mediates myotonic dystrophy kinase-related Cdc42-binding kinase (MRCK) regulation of LIMK1 protein in lamellipodial F-actin dynamics. J Biol Chem 2014; 289: 26989–27003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.