Figure 4.
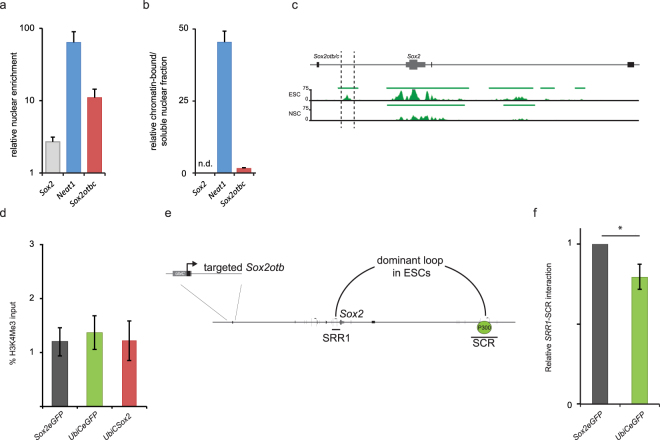
Sox2 locus-specific H3K4 trimethylation and chromatin interactions in ESCs overexpressing Sox2otb. (a) Analysis of Sox2ot RNA localization in ESCs. Sox2ot is enriched in the nucleus when compared to β-Actin as determined by qRT-PCR after subcellular fractionation. The ratio (+SD) of nuclear/total RNA (200ng input) relative to that of β-actin is depicted on a 10 log scale. Neat1 is a lncRNA that is enriched in the nucleus, and which is predominantly associated to chromatin46. (b) Analysis of the nuclear localization of Sox2ot in ESCs by qRT-PCR. The depicted ratio of chromatin bound RNA (+SD) is relative to that of β-actin. (c) Genome browser view of H3K4me3 density signals in the regulatory Sox2 region of ESCs and ESC-derived NPCs67. For quantification of the difference see Supplementary Fig. 5Sa, b, and c. (d) H3K4me3 ChIP results for the region depicted between vertical lines in (c). Depicted H3K4me3 levels are relative to H3K4me3 levels of the housekeeping gene Myl6. (e) Schematic drawing of the dominant chromatin loop in ESCs formed by interaction of the Sox2 proximal region (Sox2 regulatory region 1) (SRR1) with a P300 bound super enhancer (SCR) located ~110 kb downstream of Sox2. HindIII fragments and primers used are shown. (f) 3C chromatin conformation capture of the SRR1-SCR interaction depicted in (e). Values are relative to interactions of the Sox2 intergenic region upstream of Sox2otc. Values are represented as mean +/− SD from three independent experiments (n = 10). *Paired t-test P value = 0.02.