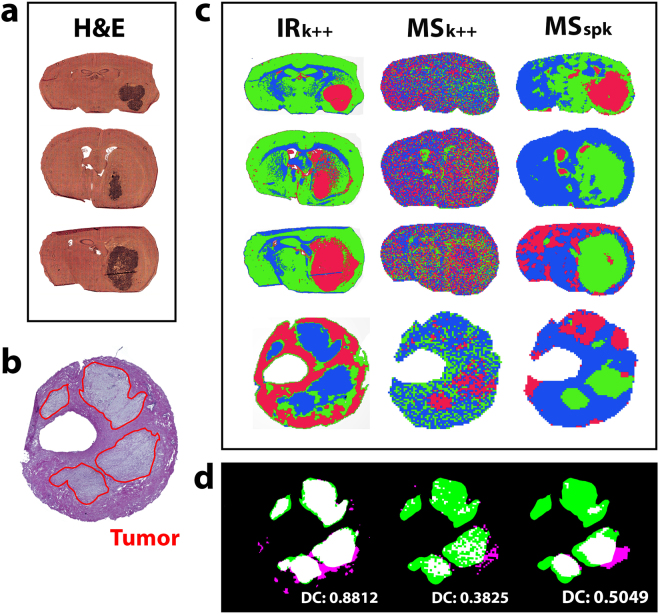
Figure 3.
Accuracy of tumor contour assignment derived from FTIR- and MS image segmentation. (a) H&E stained cryosections of CD1 nu/nu mouse brains engrafted with U87-MG glioblastoma cells on gold-coated slides and (b) H&E stains of human patient-derived GIST tissue were used to evaluate (c) IR and MS imaging based segmentation results. The images recorded from adjacent cryosections were in silico dissected into three areas of greatest possible data homogeneity, expressed by a color code (red, green, blue). The resulting segmentation images derived from FTIR images using k-means++ clustering (IRk++) and MS images using k-means++ clustering (MSk++) as well as spatially aware clustering (MSspk) are compared. (d) The spatial contours of each derived segment were automatically registered to a pathologist’s tumor annotation in human GIST tissue (red circle in b) to determine each modalities’ capability of revealing the observed tumor structure. Calculation of the DSC demonstrates a more precise (88.1% overlap between the binary tumor contours and segmentation result) disaggregation of the examined tissues when based on FTIR data compared to results based on MSI data (38.3% for MSk++ and 50.5% for MSspk).