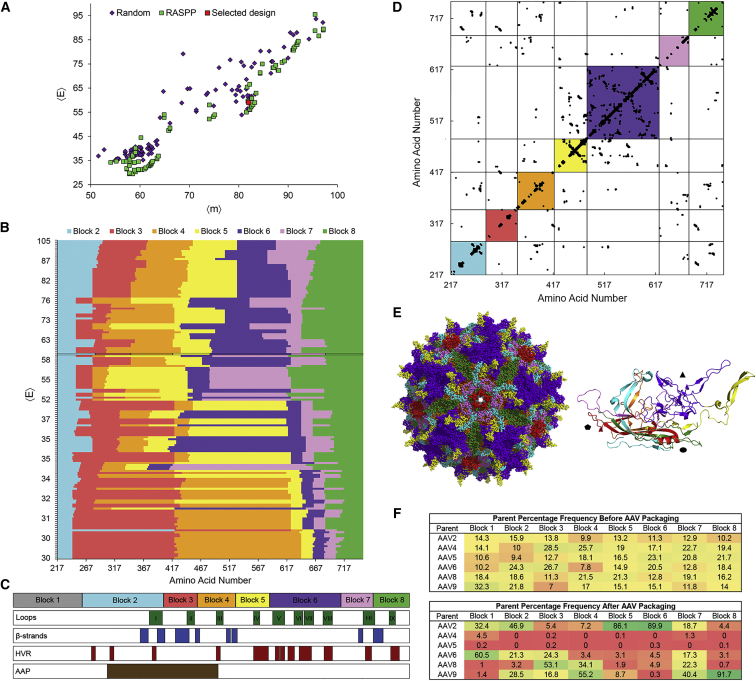
Figure 1.
SCHEMA AAV Library Design
(A) The RASPP optimization algorithm generates library designs that are lower in 〈E〉 at various mutation levels. The RASPP library design selected for construction is indicated in red. (B) RASPP library designs over a range of 〈E〉 levels. The library selected for construction is indicated by a black border. Block 1 is omitted because it lies outside of the crystallized region of the capsid for which the SCHEMA analysis can be conducted. Parameters for library design were a minimum block length of 20 amino acids and a maximum length of 250 amino acids. (C) Schematic of cap crossover positions in the AAV library design selected for construction. An alignment of capsid loops, β strands that form the anti-parallel β-barrel motif, hypervariable regions (HVR), and the assembly-activating protein (AAP) are provided to indicate known structure-function relationships in the AAV capsid. (D) Protein contact map of the selected library design. All possible residue-residue contacts are displayed as black dots. The colored squares represent the sequence blocks that are shuffled. Contacts retained within the colored squares are preserved during recombination, while contacts outside of these squares may be broken depending on the identity of the parent sequences at each block. (E) Three-dimensional models of the selected capsid design. The shuffled blocks are represented by the corresponding colors used in (B)–(D) and mapped onto the AAV2 crystal structure (PDB: 1LP3) in PyMOL. The full biological assembly and a single asymmetric subunit with shapes indicating the axes of symmetry are shown. (F) The percentage frequency of each AAV parent before and after viral packaging of the assembled SCHEMA library are presented as a heatmap.