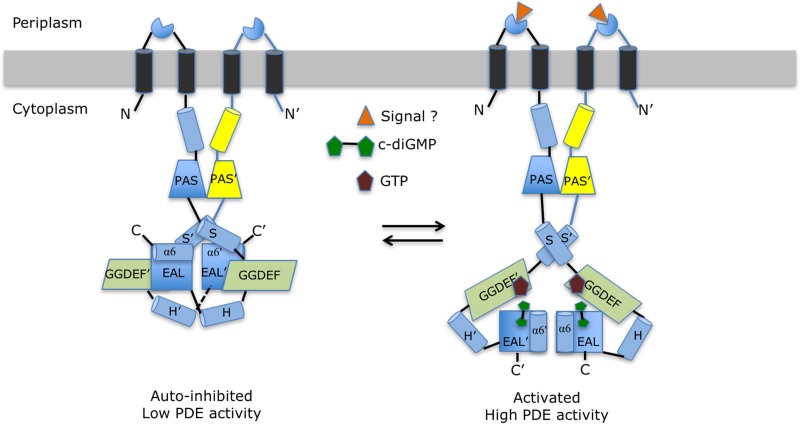
FIG 8.
Proposed allosteric mechanism regulating the PDE activity of RbdA. The left panel schematically represents the putative autoinhibited state observed in the cRbdA crystal structure. This resting state is stabilized by interactions between helix α6 from the EAL domain and the S′-helix immediately N-terminal to the GGDEF′ domain (and also between helix α6-E′ and the S-helix in the other monomer, via the dyad). These interactions lock both EAL domains of the RbdA dimer in a noncanonical configuration. Upon signal detection by either the putative periplasmic sensor domain (triangle) or GTP binding to the A sites of the GGDEF domains, local conformational changes (Fig. 5D) propagate throughout the protein and lead to the release of the autoinhibitory interactions between helix α6-E and the S′-helix. This is proposed to lead to the rearrangement of both EAL domains into a canonical dimer capable of hydrolyzing the incoming c-di-GMP substrate and to the observed higher PDE activity (Fig. 1D) conducive to biofilm dispersal (right panel).