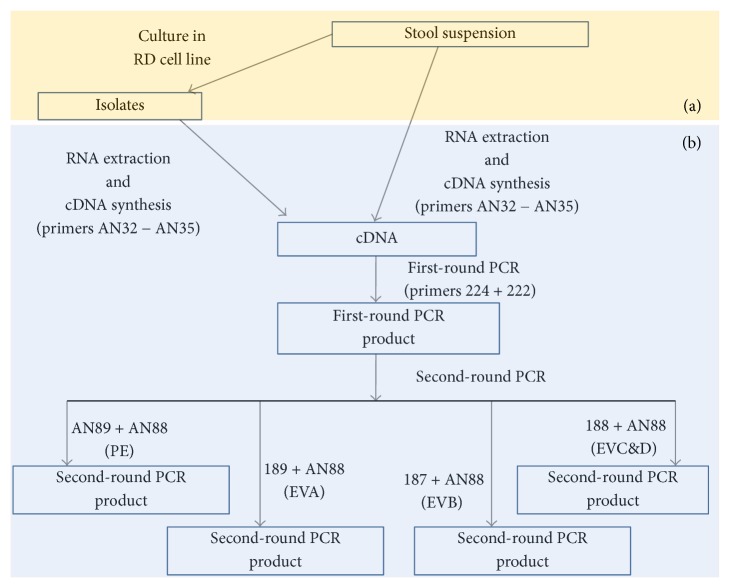
Figure 1.
Schematic representation of the algorithm used in this study. (a) Sixteen RD cell culture isolates and their corresponding sixteen fecal suspensions were collected from the WHO National Polio Laboratory in Ibadan, Nigeria. (b) RNA was extracted from all thirty-two (32) samples (RD positive isolates and their corresponding suspension) and subsequently converted to cDNA. The cDNA was used as template in the 1st round PCR assay. The first-round PCR assay product was used as template in four different second-round PCR assays. Positive samples for the 2nd round PCR assays were sequenced and the result was used for enterovirus identification.