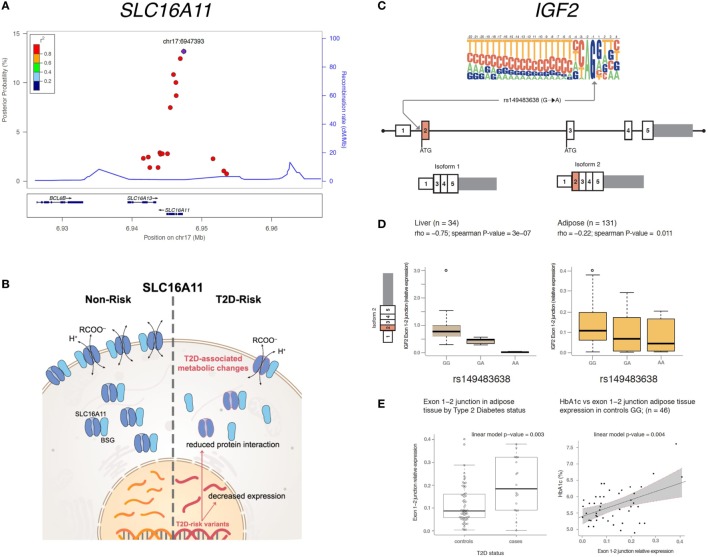
Figure 1.
Overview of genetic associations with the potential to develop into new therapeutic strategies within SLC16A11 (A,B) and IGF2 (C–E). (A) Fine mapping of the SLC16A11 region identified in Williams et al. (4) mapped and revealed several candidate variants at or near SLC16A11 gene. Each dot represents a variant within the 99% credible set, i.e,. the variants that have, in aggregate 99% probability of containing the causal variant. The y-axis represents the posterior probability of being causal, and the x-axis the genomic position (Hg19). The lead SNP is represented by the purple symbol. The color-coding scheme indicates the R-squared with the lead SNP, estimated based on the Mexican population. Only the SNPs that fall within the 99% credible set are plotted. (B) The type 2 diabetes (T2D) risk haplotype contains a cis-eQTL associated with lower SLC16A11 expression in the liver. In addition, coding risk alleles in the same haplotype disrupt the interaction between SLC16A11 and basigin (BSG). Reduced SLC16A11 expression was shown to induce metabolic changes associated with T2D. (C) rs149483638 prevents splicing in vitro and in vivo. This variant is located at a canonical splice acceptor site, and is predicted to cause skipping of exon 2 of IGF2 isoform 2. (D) The dosage of the T2D protective A allele is correlated with lower expression of IGF2 isoform 2 (as measured by expression levels of the exon 1–2 junction) in liver and in adipose tissue. (E) Expression of IGF2 isoform 2 is associated with T2D and glycated hemoglobin (HbA1c). Boxplots representing the expression of IGF2 isoform 2 across T2D cases and controls in individuals homozygous for the G common allele. The linear model P-value represents the association between IGF2 isoform 2 expression, adjusted by age, body mass index, and sex. The IGF2 isoform 2 positively correlates with higher plasma HbA1c in participants without diabetes. Figure adapted from Rusu et al. (13), Copyright 2017 by Elsevier with permission and Mercader et al. (17) American Diabetes Association and Copyright Clearance Center with permission.