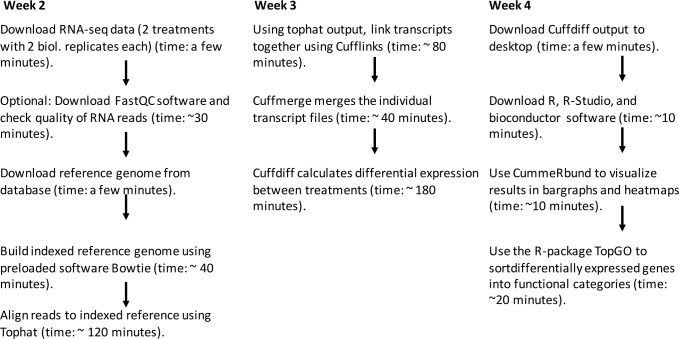
Fig 1. Flowchart depicting the compute pipeline used in the module.
In Week 1, all introductory Unix exercises are performed on the student’s computer without the need for cloud computing or a Linux cluster. In Week 1, students also sign up for a free CyVerse account and start up a virtual machine. In Weeks 2 and 3, most of the cloud computing steps are performed. The approximate compute times for each step are listed based on the use of a virtual machine with one CPU (“small” CyVerse instance). In between compute steps, students write scripts, format and verify data using command-line Unix tools, read the rationale behind each step, and spend some time watching a YouTube video (for the FastQC tutorial) and getting familiar with specific help forums for bioinformatics questions, such as Biostars. Depending on student preparedness and engagement, each lab session can be completed in 2.5 to 4 hours. Using more CPUs can speed up the compute time significantly for Tophat and CuffDiff analyses, which are in the current lab set-up “overnight” steps separating labs. CPU, central processing unit.