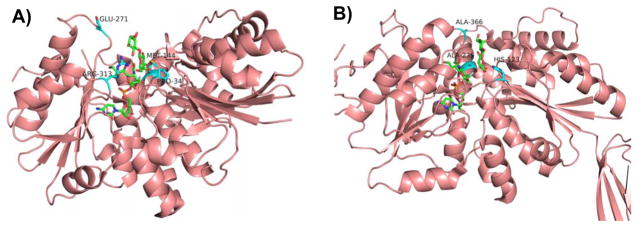
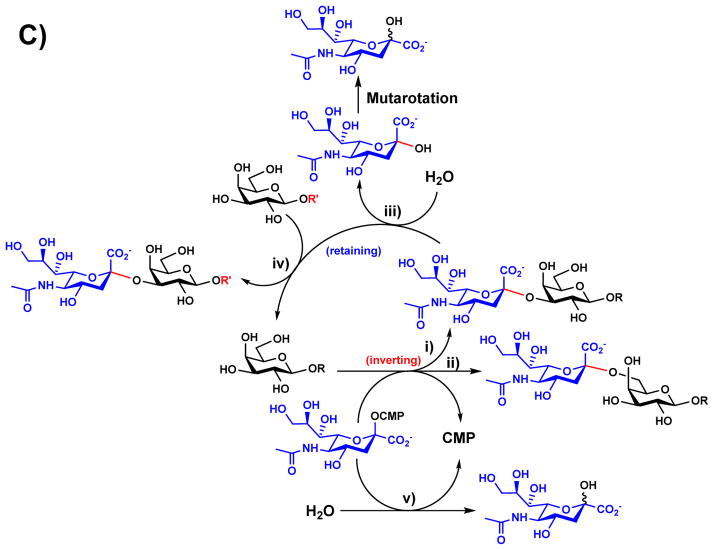
Figure 4.
Amino acid residues known to influence the activities of GT-B-fold GT80 sialyltransferases PmST1 (A) and Psp2,6ST (B) with CMP-3F-Neu5Ac and lactose as well as reactions catalyzed by multifunctional PmST1 (C). Carbon atoms in the donor and acceptor substrates are shown in green; carbon atoms in amino acid residues are shown in cyan; oxygen atoms are shown in red; and nitrogen atoms are shown in blue. Images are constructed in PyMOL from template PDB structures 2IHZ for PmST1 and 2Z4T for Psp2,6ST. The image of the Psp2,6ST structure depicts ligands from 2IHZ within the aligned 2Z4T structure.