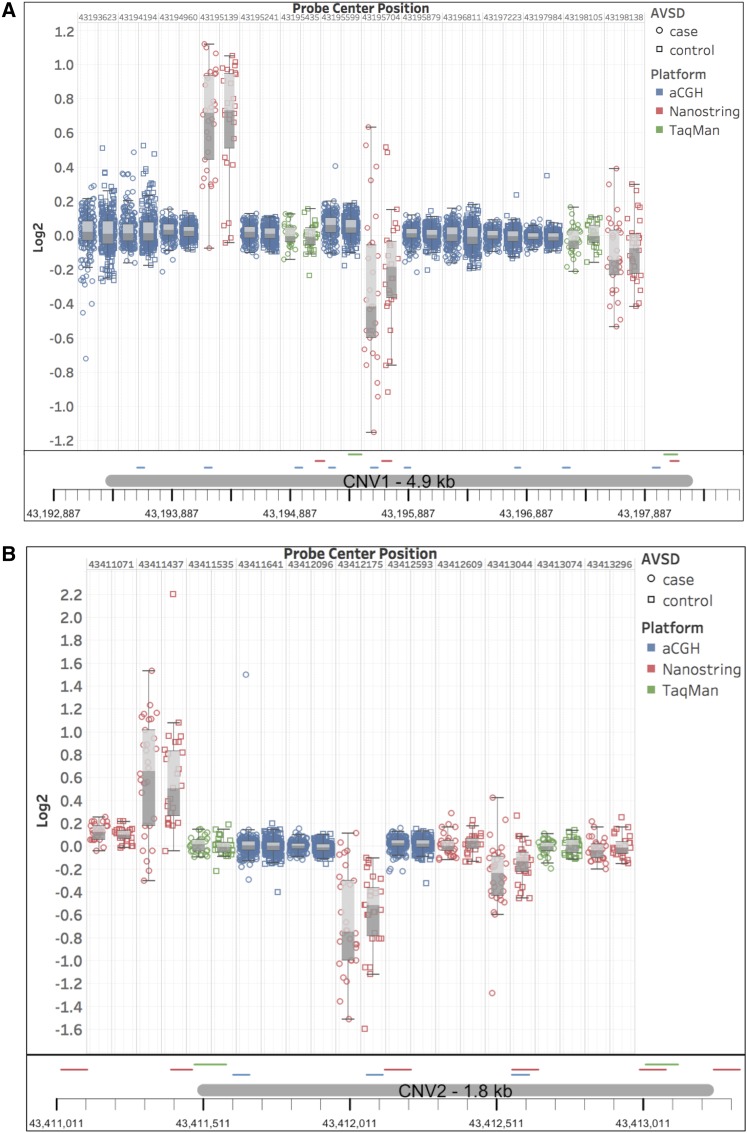
Figure 1.
Summary results from analyses to attempt to replicate previously reported DS+AVSD (Down Syndrome with a complete atrioventricular septal defect)-associated common copy number variants (CNVs) with three technologies. (A and B) shows measures for Sailani-putative CNVs 1 and 2, respectively. Boxplots show inner quartile range of log2 ratios for tested DS samples with whiskers reaching 1.5 times the interquartile range. Case values are circles, and control values are in outlined boxes. Array comparative genomic hybridization (aCGH) probes are blue, NanoString probes are red, and TaqMan probes are green. Boxplots are ordered by genomic location, and their precise locations are indicated beneath the plots. Neither CGH probes nor TaqMan Copy Number assays detected aberrant copy numbers or differences between cases and controls. Varying results were found across these loci by NanoString probes, with some probes showing differences in log2 means between cases and controls. Within these same proposed small CNV loci, NanoString probes called all possible combinations of copy gain, loss, and no change within the same small cohort. When compared to adjacent CGH and TaqMan probes, it is clear that the NanoString probes are not reliable predictors of copy number state at this locus.