Figure 4.
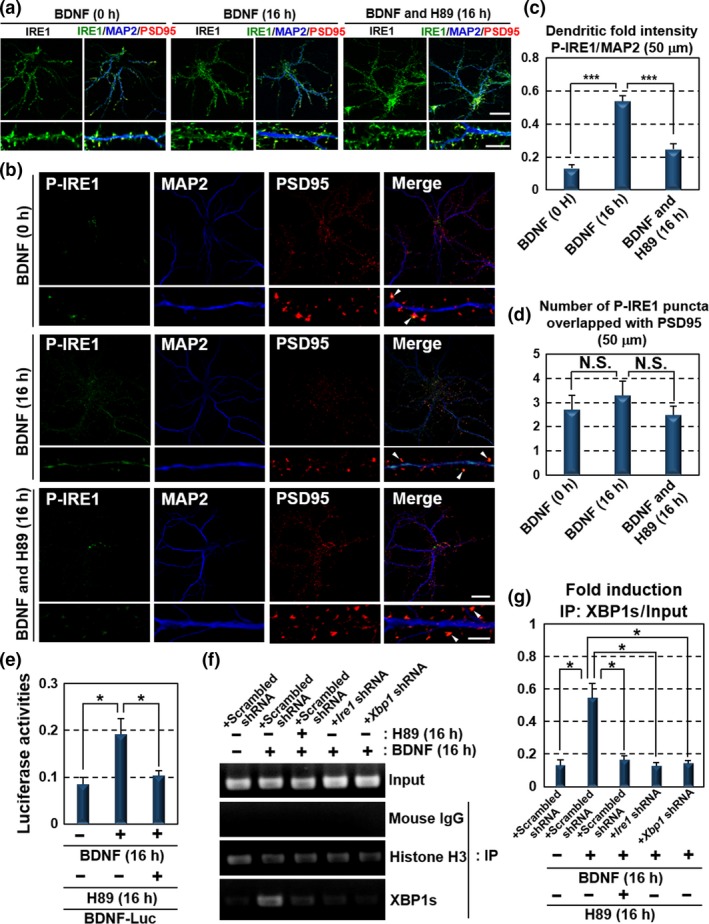
BDNF drives its own expression through dendritic activation of the PKA‐IRE1‐XBP1 pathway. (a) Immunofluorescence staining analysis of IRE1 (green), MAP2 (blue) and PSD95 (red) in primary cultured hippocampal neurons maintained for 10 days. The localization and level of IRE1 were not significantly changed by the exposure to 100 ng/mL BDNF or 30 μM H89 for 16 h. The lower panels show high magnification of the dendrites shown in the upper panels (Bar in upper panels: 20 μm, in lower panels: 5 μm). (b) Immunofluorescence staining analysis of P‐IRE1 (green), MAP2 (blue) and PSD95 (red) in primary cultured hippocampal neurons maintained for 10 days. IRE1 was phosphorylated at MAP2‐positive dendrites following exposure to 100 ng/mL BDNF for 16 h. In contrast, phosphorylation level of IRE1 in PSD95‐positive postsynaptic sites was decreased compared with those of the glutamate treatment presented in Fig. 1(e). BDNF‐induced IRE1 phosphorylation was reduced by treatment with 30 μM H89 for 16 h. The lower panels show high magnification of the dendrites shown in the upper panels. Arrow heads indicate immunoreactivities of P‐IRE1 overlapped with those of PSD95. (Bar in upper panels: 20 μm, in lower panels: 5 μm). (c) P‐IRE1 fluorescence intensities 40–90 μm from the somata of the dendrites in (b). The fluorescence intensities in these dendrites were increased by BDNF treatment (mean ± SD, N = 10; number of cells prepared from five independent cultures; ***p < 0.001). (d) The number of P‐IRE1‐positive puncta overlapping with PSD95‐positive postsynaptic sites 40–90 μm from the somata in (b). The number of P‐IRE1‐positive postsynaptic sites was not significantly increased by BDNF exposure (mean ± SD, N = 10; number of cells prepared from five independent cultures). (e) Reporter assays using Neuro‐2A cells. Cells transfected with the BDNF‐Luc constructs shown in Fig. 2(d) were treated with 100 ng/mL BDNF and 30 μM H89 for 16 h. Reporter activities induced by BDNF were inhibited by H89 treatment (mean ± SD, N = 6; number of samples prepared from independent cultures; *p < 0.05). (f) ChIP assay using primary cultured hippocampal neurons maintained for 10 days. Cells were infected with lentiviruses expressing scrambled, Ire1 or Xbp1 shRNA, with GFP for 5 days, and treated with 100 ng/mL BDNF and 30 μM H89 for 16 h. The primer set used for PCR amplification is shown in Fig. 2(f). Chromatin immunoprecipitation was performed using the indicated antibodies and mouse IgG (negative control). (g) Quantitative analysis of PCR amplification after immunoprecipitation by anti‐XBP1s antibody in (f) (mean ± SD, N = 3; number of samples prepared from independent cultures; *p < 0.05).
