Fig. 1.
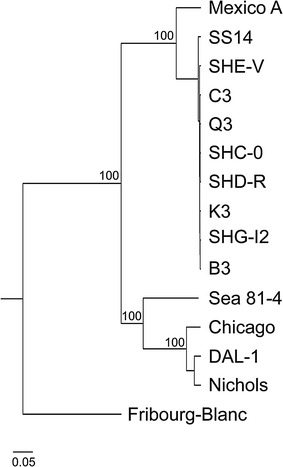
The phylogenetic tree of Treponema genomes—reposition of Chinese samples. The phylogenetic tree was constructed using hierarchical clustering based on multiscale bootstrap resampling. The number of bootstrap replications was set to 1000. The bar scale represents the number of substitutions per site. The analysis involved 14 TPA genome sequences including eight derived from the Chinese samples: SHC-0, SHD-R, SHE-V, SHG-I2, B3, C3, K3, and Q3. The T. pallidum subsp. pertenue Fribourg-Blanc genome sequence was used as an outgroup. There was a total of 2444 SNV positions in the final dataset (see Additional file 9). Two separate clusters supported by a bootstrap confidence level greater than 95% were identified: one cluster consisted of Nichols-like TPA strains (TPA Lineage 1), the other consisted of SS14-like TPA strains including all tested Chinese strains (TPA Lineage 3). A total of 1,130,357 nucleotide positions were aligned in the final dataset
