Figure 1. Maturation kinetics and its impact on fluorescence signal in growing cells.
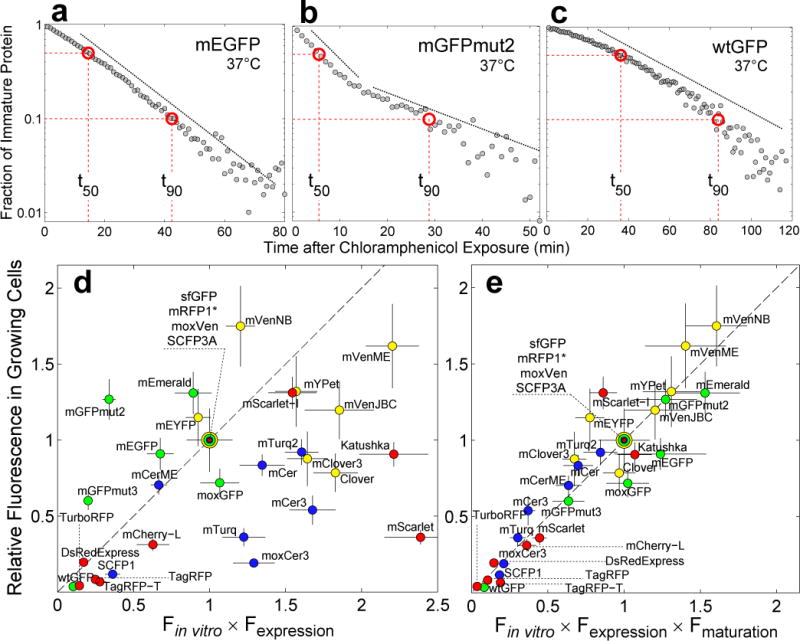
(a), (b) and (c). Fraction of immature protein as a function of time after translational arrest with chloramphenicol in single cells. (a) mEGFP maturation kinetics shows a single exponential decay (dashed line). (b) mGFPmut2 exhibits a more complex maturation process with two kinetic steps (dashed lines). (c) wtGFP matures with a slow non-exponential rate that increases with time. Dashed red lines indicate the time it takes for 50%, t50, and 90%, t90, of FP to mature, respectively. (d) Fluorescence signal in growing cells vs Fin vitro=QY·ε multiplied by Fexpression=amount of protein. Fluorescence signal is the mean fluorescence of single-cells (exponential growth in single-cell chemostat, M9 rich media, 37°C, mean from 70±20 cells, ±SEM). QY and ε are the values reported when the FPs were first published except for mVenusME and mCeruelanME; for these two FPs, we used our own in vitro data, see Online Methods. Fexpression was estimated using SDS-PAGE gel densitometry (±SD, see Online Methods). For green FPs, data normalized by sfGFP data; for yellow FPs, by moxVenus data; for blue FPs, by SCFP3A data; and for red FPs by mRFP1*. Dotted line is the identity. Dilution is given by the doubling time of E. coli, tgr=28.5±2min. (e) Fluorescence signal in growing cells vs Fin vitro × Fexpression multiplied by Fmat=1/(1 + t50/tgr)19. Fluorescence signal data is the same as in (d). QY and ε were quantified in our laboratory independently, except for in vitro data of red FPs, see Online Methods. We have assumed that the in vitro brightness of mRFP1* and mCherry-L is the same as the in vitro brightness of mRFP1 and mCherry, respectively, because amino acid differences are not part of the β-barrel. Error bars calculated by propagation of QY, ε and t50 errors.
