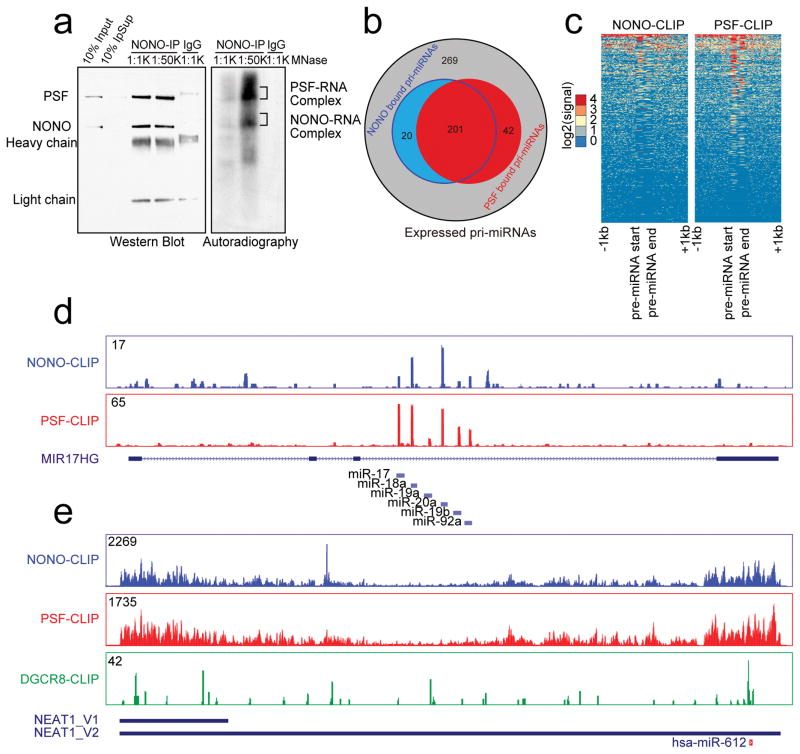
Fig. 3. Genome-wide analysis of NONO-PSF-RNA interactions.
(a) Immunoprecipitated NONO-PSF crosslinked to RNA. The complex was treated with two different concentrations of MNase (1:1,000 or 1:50,000 dilution); RNA in the complex was 32p-labeled with T4 polynucleotide kinase. Proteins and RNA were visualized by Western blotting (left) and autoradiography (right). Indicated bands (right) were individually isolated for CLIP-seq library construction. (b) Venn Diagram showing overlapped pri-miRNAs bound by NONO and PSF in HeLa cells. (c) Footprint of NONO and PSF on pri-miRNAs. (d) Representative NONO and PSF binding tracks on the pri-miR-17–92a transcript. (e) The binding profiles of NONO and PSF on NEAT1 in comparison with the published DGCR8 CLIP-seq signals44. Y-axis in d and e shows CLIP-seq read density in each case. The region encoding for miR-612 is indicated at bottom. Uncropped images of Western blots and autoradiography in a are shown in Supplementary Data Set 1.