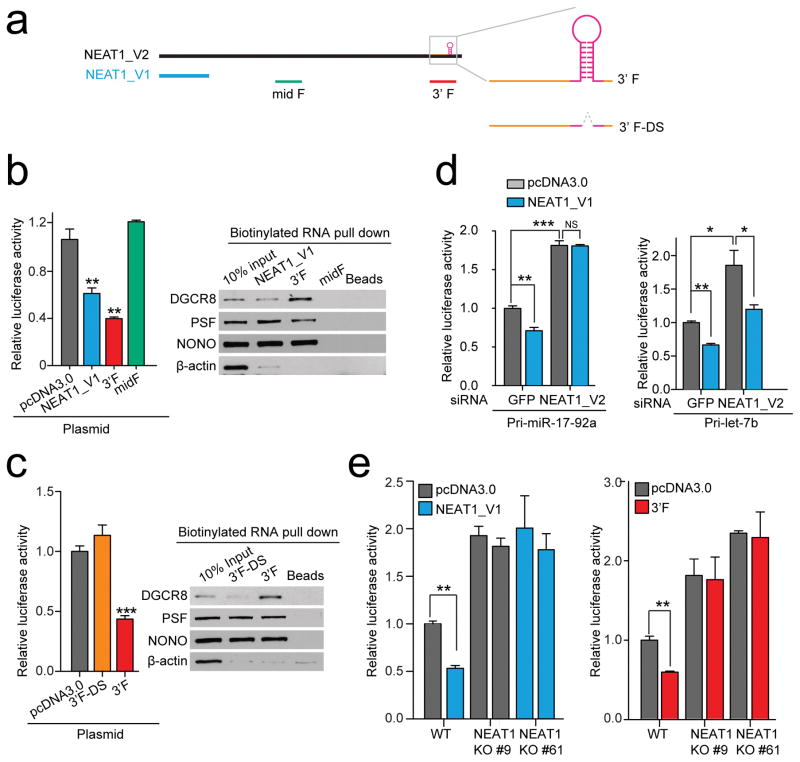
Fig. 5. NEAT1-mediated interaction networks for enhancing pri-miRNA processing.
(a) Illustration of NEAT1_V1, NEAT1_V2 and derived RNA fragments. Highlighted on the right are pri-miR-612 near the 3′ end of NEAT1_V2 and the 3′ fragments before (3′F) or after deletion of the pre-miR-612 stem-loop (3′F-DS). (b, c) Enhanced processing of the pri-miR-17–92a reporter by NEAT_V1 and 3′F, but not a middle fragment from NEAT1_V2 (midF) or 3′F-DS (left panels). The right panels show RNA pulldown results from HeLa nuclear extracts, analyzed by Western blotting for NONO-PSF and DGCR8. (d) Knockdown of NEAT1_V2 diminished the enhancement of pri-miRNA processing by overexpressed NEAT_V1 on the pri-miR-17–92a (left) or pri-Let-7b (right) processing reporter. (e) Knockout of NEAT1_V2 prevented the enhancement of pri-miRNA processing by overexpressed NEAT_V1 and the 3′F on the pri-miR-17–92a processing reporter. Bar graphs in b,c,d, and e are presented as mean ± SEM (n=3, cell culture). *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant, determined by two-tailed Student’s t test. Uncropped images of Western blots in b and c are shown in Supplementary Data Set 1 and data source for the bar graphs are reported in Source Data for Figure 5.