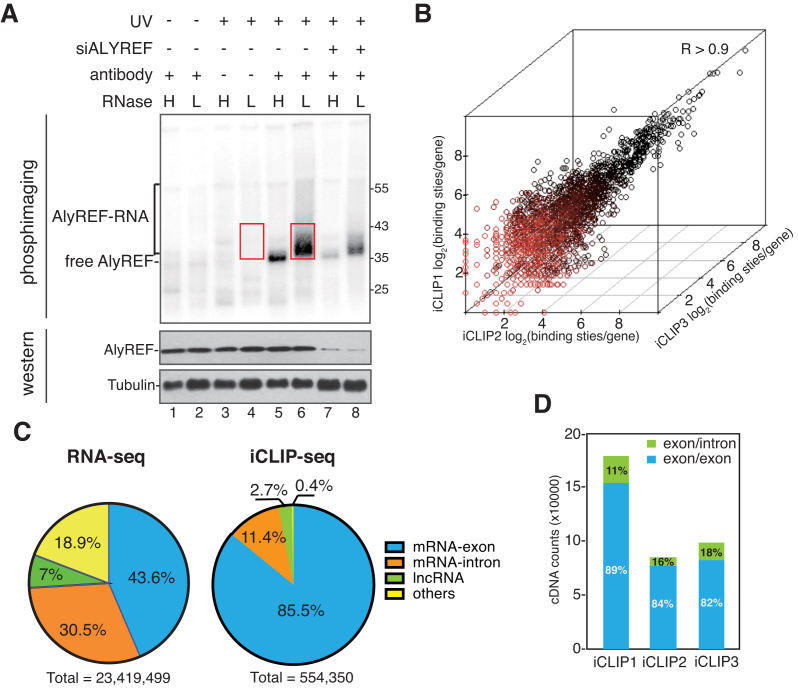
Figure 1.
ALYREF binding sites are enriched on exons of mature mRNAs. (A) Detection of ALYREF-RNA complexes. RNase I treated (H: 5 U/ml; L: 0.5 U/ml) and 32P-labeled RNP complexes were immunoprecipitated with or without the ALYREF antibody from normal and ALYREF knockdown cells. After size-separation using denaturing gel electrophoresis, ALYREF-RNA complexes were transferred to a nitrocellulose membrane. The upper panel shows the autoradiograph of the nitrocellulose membrane. The lower panel shows the western blotting result using the indicated antibodies for input of IP. Red box indicates the region that was extracted for subsequent analyses. (B) Correlation of ALYREF iCLIP-seq replicates. Clustered ALYREF binding sites in each gene are plotted for three independent biological replicates (Spearman correlation coefficient, R > 0.90 for all comparisons). (C) ALYREF binding sites are enriched at mRNA exon. Left pie chart shows the percentage of RNA-seq reads that uniquely mapped to four human genome regions: mRNA exons, mRNA-introns, lncRNAs or others. Right pie chart shows the percentage of clustered ALYREF binding sites mapped to different human genome regions. (D) ALYREF mainly binds mature mRNAs. Percentages of the iCLIP tag in ALYREF binding sites specifically mapped to the exon-exon junction (blue) or exon-intron junction (green).