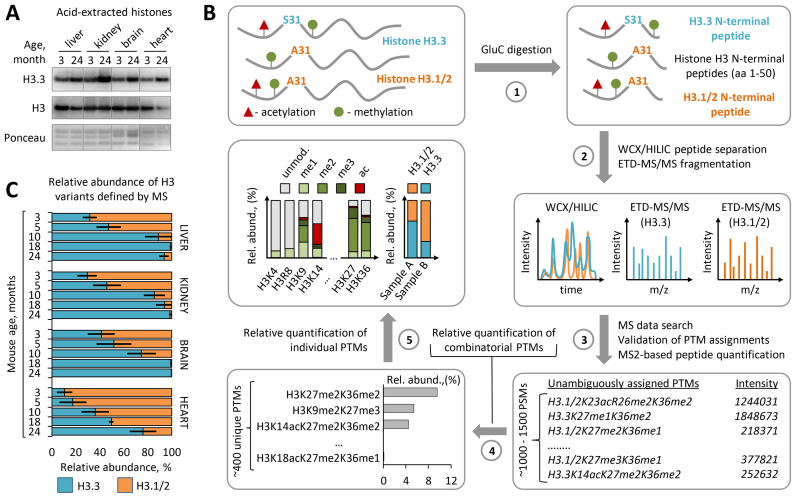
Figure 1.
Histone H3.3 accumulates and replaces canonical H3 isoforms with age. (A) Immunoblot analysis of H3.3 levels in histone extracts from somatic tissues of 3-month-old and 24-month-old C57BL/6J mice. Total histone H3 and Ponceau S staining were used as loading controls. (B) Overview of the middle-down proteomics workflow used in the study. Histones were acid extracted from mouse tissues and digested with endopeptidase GluС (1). The resulting peptide mixtures containing 50 aa residues long H3 N-terminal peptides were analyzed by on-line nano-LC–MS/MS method using weak cation exchange–hydrophilic interaction liquid chromatography (WCX-HILIC) for peptide separation and electron transfer dissociation (ETD) for their fragmentation (2). Acquired MS/MS data were searched against a histone protein database to obtain a set of peptide identifications followed by filtering out of hits with ambiguous PTM site assignments (3). Peptides with unambiguous PTM localizations were quantified based on the total ion current of their MS/MS spectra (MS2-TIC). Obtained quantitative results were used to calculate the relative abundances of distinct H3 proteoforms barring single or combinatorial PTMs, where the sum of all MS2-TIC intensities was considered as 100% (4). The relative abundance of each individual H3 PTM and each H3 variant was calculated by summing up the relative abundances of all H3 proteoforms containing the given PTM or assigned to the given H3 variant, respectively (5). The relative abundance of individual PTMs on H3.3 was calculated as the sum of the relative abundances of H3.3 proteoforms containing the given PTM normalized by the relative abundance of the H3.3 variant. For more details, see the ‘Materials and Methods’ section. (C) Dynamics of H3.3 accumulation in chromatin of various mouse somatic tissues with age as revealed by middle-down MS analysis. Error bars indicate SD (n = 2–4 biological replicates).