Abstract
Splicing is initiated by a productive interaction between the pre-mRNA and the U1 snRNP, in which a short RNA duplex is established between the 5′ splice site of a pre-mRNA and the 5′ end of the U1 snRNA. A long-standing puzzle has been why the AU dincucleotide at the 5′-end of the U1 snRNA is highly conserved, despite the absence of an apparent role in the formation of the duplex. To explore this conundrum, we varied this AU dinucleotide into all possible permutations and analyzed the resulting molecular consequences. This led to the unexpected findings that the AU dinucleotide dictates the optimal binding of cap-binding complex (CBC) to the 5′ end of the nascent U1 snRNA, which ultimately influences the utilization of U1 snRNP in splicing. Our data also provide a structural interpretation as to why the AU dinucleotide is conserved during evolution.
INTRODUCTION
Nuclear pre-mRNA splicing is an evolutionarily conserved process that removes introns from precursor messenger RNAs (pre-mRNAs) to produce mature mRNAs. Intron removal is executed by the spliceosome, a large macromolecular machine consisting of five small nuclear ribonucleoprotein particles (snRNPs), U1, U2, U4, U5 and U6 (1–3). Each of these snRNPs contains one small nuclear RNA (snRNA) together with a set of proteins (1–4). Unlike other macromolecular machines, such as the ribosome and RNA polymerase II (pol II), the spliceosome is assembled de novo on pre-mRNA substrates in an ordered and dynamic fashion through sequential binding of snRNPs and wholesale exchanges of splicing factors (2). The U1 snRNP first binds to the intron 5′ splice site (5′ss), which is followed by U2 snRNP recruitment to the intron branch site and the subsequent joining of U5-U4/U6 tri-snRNP to form a pre-catalytic spliceosome, which then progresses through a series of highly coordinated dynamic remodeling steps to produce a mature spliceosome that is activated for chemical catalysis (1,2,5).
During spliceosome assembly, productive binding of the U1 snRNP to the 5′ss is considered one of the most critical steps, because it commits a pre-mRNA to the splicing pathway (6–10). The successfully formed U1-snRNP-containing complexes are called commitment complexes (CCs). In the budding yeast, two commitment complexes, CC1 and CC2, can be detected by native gel electrophoresis in vitro (6,7). The formation of CC1 requires a functional 5′ss, whereas the subsequent formation of CC2 additionally requires a functional branch site, as well as branch-site-binding protein (BBP) and Mud2p, the yeast orthologs of mammalian SF1 and U2AF65, respectively (11–14). Several U1-snRNP proteins and other non-snRNP proteins also make energetic contributions to the CC formation and stability (15–23), including the heterodimeric cap-binding complex (CBC) consisting of Cbp20p and Cbp80p (24–26).
Several lines of evidence suggest that the CBC is functionally involved in splicing, distinct from its well-established role in binding to the 7-methylguanosine (m7G) cap structures that are co-transcriptionally added to pol II transcripts in the nucleus. First, depletion of Cbp20p markedly reduces CC formation and splicing efficiency in vitro (27–30). Second, genetic perturbations of the CBC result in inefficient splicing in vivo (31,32). Third, the CBC is genetically linked to BBP, Mud2p, and several U1-snRNP components (31–33). However, understanding the precise role of the CBC in splicing is complicated by the fact that it can also bind to the m7G cap of the nascent pol II-synthesized snRNAs in the nucleus. In mammalian cells, biogenesis of these snRNPs involves export of the partially assembled complex to the cytoplasm (1,34,35), where the m7G is converted into a trimethylguanosine (TMG) structure through the activity of trimethyl guanosine synthase (Tgs1p in budding yeast) (36–38). The TMG-capped snRNPs are then re-imported into the nucleus for their respective functions in splicing (1,35,39,40). Because the CBC preferentially binds m7G over TMG (34,41), the cytoplasmic hypermethylation reaction presumably reduces the affinity of the CBC for the mature snRNPs. This hypothesis is supported by the findings that the CBC co-purifies with the U1 snRNP from the tgs1Δ cells (42) and that the cold-sensitive growth defect of tgs1Δ cells can be rescued by mutations in CBP20 that are specifically designed to relax the physical contacts between Cbp20p and the m7G cap (42,43). Thus, it appears that the correct methylation status enforced by Tgs1p and possibly other characteristics of the 5′ cap dictates the CBC occupancy, which in turn impacts the ultimate functionality of at least the U1 snRNP (38,42,43).
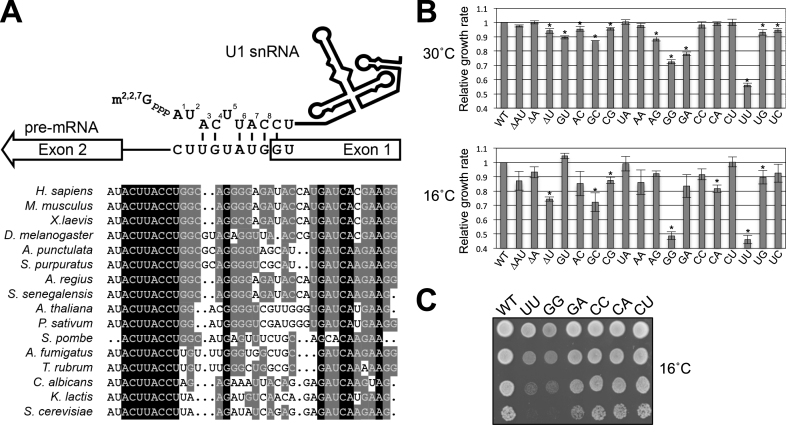
It is well established that the 5′ end of the U1 snRNA (positions 3–8) forms specific basepairs with the 5′ss of the pre-mRNA, which in budding yeast introns generally conform to a nearly invariant consensus sequence, GUAUGU (44,45) (Figure 1A). Although seemingly thermodynamically weak, mutational and suppression analyses demonstrate that the strength of this 5–7 bp RNA duplex is the primary determinant of U1 snRNP recognition and selection of authentic 5′ss during splicing (46–49). These studies did not, however, address the impact of the potential interaction between the first two nucleotides of U1 snRNA (i.e. AU) and positions 7–8 of the 5′ss region, which show no apparent sequence conservation (44,45). Interestingly, artificially introducing basepairing between the AU dinucleotide of U1 snRNA and intron positions 7–8, which hyperstabilizes the U1 snRNA/5′ss interaction, resulted in either reduced (50) or enhanced (51) splicing efficiency. Recent in vivo studies in the mammalian system further cloud the picture by showing that U1 snRNA/5′ss helices may accommodate noncanonical registers such as bulged duplexes (52–54), thereby rendering it difficult to predict the outcome of extending the basepairing through the AU dinucleotide. These data therefore make it even more puzzling as to why the AU dinucleotide at the 5′ end of U1 snRNA is highly conserved, except in Schizosacchromyces species (55) (Figure 1A and Supplementary Figure S1).
Figure 1.
Alteration of U1 snRNA 5′-end AU dinucleotide results in growth defect. (A) Base-pairing interaction between U1 snRNA 5′ end and the 5′ss. U1 snRNA is proposed to form a 6-bp interaction with the canonical 5′ss (GUAUGU). The 5′-end AU dinucleotide, although highly conserved (bottom panel), is not predicted to universally interact with 5′ss. Note that the m7G cap is co-transcriptionally added to the 5′ end of U1 snRNA and further modified into m2,2,7G cap. The AU dinucleotide is uniquely missing in S. pombe. (B) The growth rates of yeast strains producing U1 snRNA variants relative to an isogenic wild-type strain were determined by a competitive fitness assay. Strains exhibiting significantly low fitness scores (P < 0.01, two-tailed t-test) at 30°C or 16°C were indicated (*). Data represent the mean ± S.E.M. of three independent biological replicates. (C) Growth phenotypes of strains expressing U1 snRNA variants. Relevant genotypes of the yeast strains are shown on the left of the panel. Cells were grown to saturation at 30°C, serially diluted, and spotted on YPD plates for incubation at 16°C.
To explore the potential role of the conserved AU dinucleotide at the 5′ end of U1 snRNA, we systematically altered the dinucleotide into all possible combinations. Through a combined classical and genome-wide genetic analysis as well as detailed biochemical probing, we showed that the AU dinucleotide identity is critical for the optimal interaction of CBC with the 5′ end of the U1 snRNA, which ultimately impacts its functionality in splicing at the CC formation step.
MATERIALS AND METHODS
Competitive fitness assay
The U1-snRNA-variant strains used in this assay were all derived from YTC1585 (MATasnr19::LYS2 cup1::ura3 his3 leu2 ade2 trp1 pSNR19001 [SNR19/LEU2/CEN]) (50) via plasmid shuffling. In that, pSNR19001 was individually replaced by a variant of pSNR19001 expressing a specific version of U1 snRNA altered at its 5′ dinucleotide by standard oligonucleotide site-specific mutagenesis. Fitness of an U1-snRNA variant strain was quantitatively measured by placing the query strain in direct competition with a GFP-expressing reference strain (56). In brief, an equal number (104) of early-log-phase query and reference cells were placed in the same culture for growth at the desired temperature for 19 h. A fluorescence-activated cell sorter (BD FACSCalibur™) was then used to count the number of GFP-expressing cells for a total number of 1000 cells in the resulting cell mixture. The time-zero control was also counted in the same manner.
Primer extension analysis
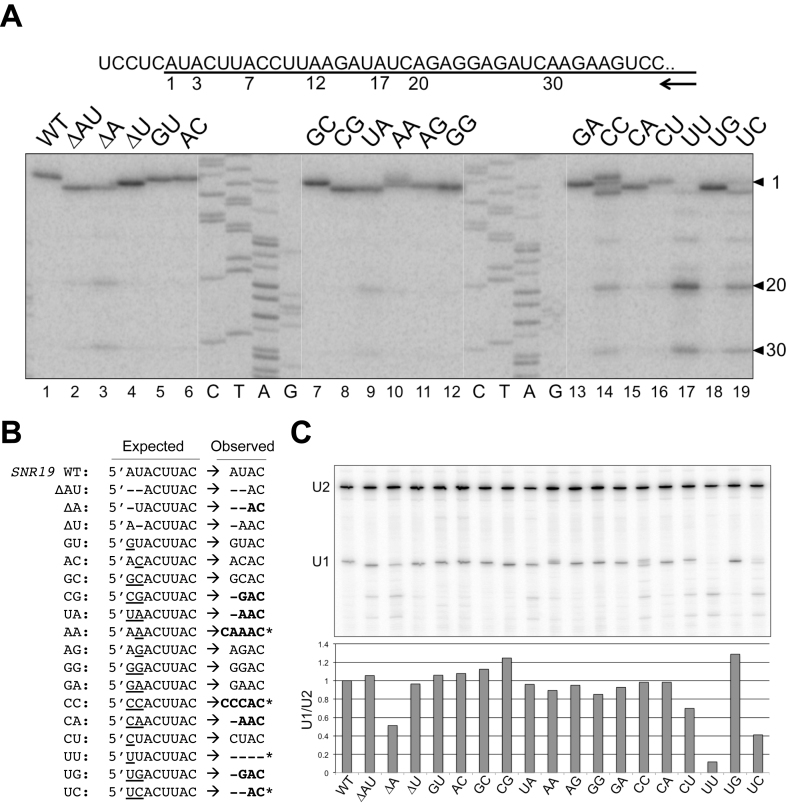
Radioactively end-labeled oligonucleotide (∼5.8 × 104 cpm) was used as primer in a 20-μl reverse transcription reaction that contained 50 μg of yeast total RNA at 45°C for 1 h (57). DNA products were recovered after degrading the RNA by NaOH and fractionated on a 6% sequencing gel (acrylamide-bisacrylamide [19:1]/8 M urea). The same primer was used to generate the sequencing ladder using pSNR19001 as a template. Sequencing reaction was done by using Sequenase™ Version 2.0 DNA Sequencing Kit (Affymetrix). Individual band intensities shown in Figure 2 were quantitated by ImageJ (https://imagej.nih.gov/ij).
Figure 2.
Alteration of U1 snRNA 5′-end dinucleotide impacts U1 snRNA transcription. (A) Transcription start sites of the U1 snRNA variants were mapped by primer extension. Sequence around the U1 snRNA transcription start is shown in the top panel, with the U1 snRNA sequence underlined. The numbers (1, 3, 7, 12, 17, 20 and 30) below the line indicate the wild-type and variant U1 snRNA start sites determined. The arrow represents the oligonucleotide primer used in the reverse transcription reactions, which is complementary to U1 snRNA positions 266–285. Image of the primer extension results are shown in the bottom panel. The sequencing ladder (C, T, A, G) was generated by Sanger sequencing off a plasmid harboring SNR19 using the same primer. (B) Summary of the transcription start sites used by U1 snRNA variants. Deletions are marked as short dash lines and base substitutions are underlined. Variants that have unexpected start sites are shown in bold, and those which have multiple strong start sites are marked by stars. For example, AA and CC variants have a significant start from the -1 position. We noted the U1 snRNA transcription appears to prefer A and G, although there are three cases of C start (AA, CC, and CU mutants). (C) Quantitation of the full-length U1 snRNA levels in U1 snRNA mutants by primer extension normalized to the U2 snRNA levels.
Splicing-sensitive microarrays analysis
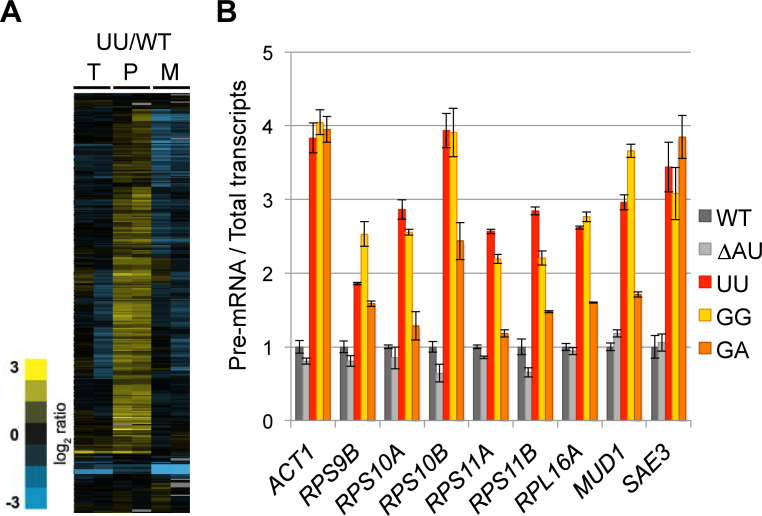
Sample collection and splicing microarray analysis were conducted as described previously (58,59). In brief, total RNAs extracted from wild-type and UU mutant strains were converted into Cy3- and Cy5-labeled cDNAs, respectively, by random priming. These samples were then hybridized to microarrays to obtain raw data for processing into three datasets (P, pre-mRNA; M, mRNA; and T, total transcripts) that enumerate the differential hybridization results of the 249 intron-containing genes. Dye-flipped replicates were conducted in each individual microarray. The splicing microarray data have been deposited to GEO (GSE97015).
Analysis of splicing efficiency by RT-qPCR
Yeast strains expressing various U1 snRNA variants were used for extracting total RNAs by standard hot phenol/chloroform method. RNA samples were converted into cDNA by random priming following manufacturer's instructions (RevertAid H Minus First Strand cDNA synthesis kit, ThermoFisher Scientific). We used Primer Express software (ver. 3.0.1, Applied Biosystems) to design all the primer pairs (Supplementary Table S1) for qPCR reactions, which were done by using StepOnePlu™ Real-Time PCR System with Fast SYBR® Green Master Mix. Data acquisition and processing were done by StepOne Software (v. 2.3; Applied Biosystems). We routinely determined the amplification efficiency of each primer pairs for normalizing the data between different primer pairs. To reduce pipetting errors, automated pipetting system (EzMate 401, Arise Biotech) was used. To measure splicing efficiency of a selected intron-contain gene, two primer sets were used to separately amplify an intron region and an exon 2 region, corresponding respectively to the levels of pre-mRNA (P) and total (T) transcripts. The ratio of P/T in the mutant background was then normalized against the same ratio obtained from the wild-type strain. All data were averages of at least three biological repeats.
Analysis of U1 snRNP and CCs by native gel electrophoresis
Active yeast splicing extracts were prepared according to the standard dounce protocol (60,61) or to the liquid-nitrogen grinding method (60,62). The procedure for probing U1 snRNP on the native gel has been previously described in detail (19). For CC detection we adapted a previously published protocol (7). Briefly, a 10-μl splicing reaction containing splicing extract (40%), an U2-specific DNA oligonucleotide RB60 (16.7 ng/μl) (63), and [32P]-labeled RP51A transcript (103 cpm/μl) was assembled in the absence of ATP and incubated at 25°C for 30 min. The reaction mixture was then mixed with equal volume of ice-cold RY buffer (50 mM HEPES [pH 7.5], 2 mM magnesium acetate, 20 mM EDTA, 1 μg/ml of total yeast RNA, and 1 mg/ml of heparin) and incubated on ice for 10 min prior to native gel electrophoresis.
Chromatin immunoprecipitation (ChIP) analysis
ChIP analysis was done exactly as described (19), except that anti-Prp40p was used to precipitate the chromatin-bound U1 snRNP. Six primer sets were used to amplify different regions of the ACT1 gene (Supplementary Table S1). The enrichment of Prp40p-bound chromatin over background was normalized to that of primer set 1 data points. Data from six biological repeats were averaged and shown in Figure 4B.
Figure 4.
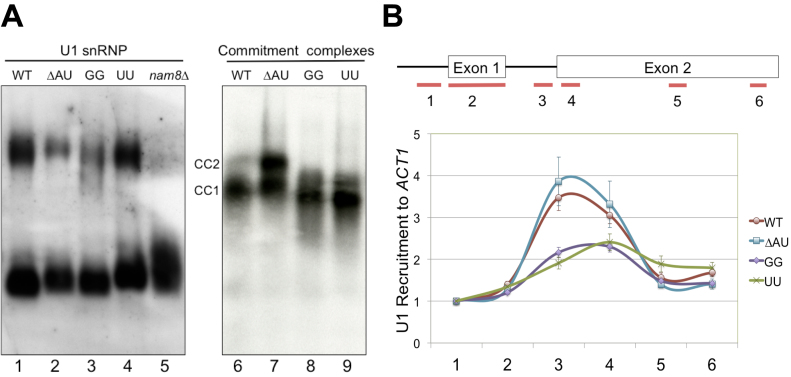
Alteration of U1 snRNA 5′-end dinucleotide impacts on U1 snRNP recruitment to pre-mRNA. (A) Native gel electrophoresis analysis of U1 snRNP and commitment complexes. (Left Panel) Alteration of U1 snRNA 5′-end dinucleotide does not appear to affect U1 snRNP mobility on the native gel. Splicing extracts made from wild-type, ΔAU, UU, GG and nam8Δ strains were electrophoresed on a native polyacrylamide (3%)–agarose (0.5%) gel. The RNAs were transferred to a membrane and probed by a digoxigenin-labeled U1 probe. No apparent differences were detected for the two major forms of the U1 snRNP except for nam8Δ. (Right Panel) Electrophoretic mobilities of CCs are altered in the presence of GG and UU mutations. CC1 and CC2 are two commitment complexes formed in the absence of U2 snRNP with [32P]-labeled RP51A transcript. (B) The GG and UU mutations severely reduce U1 snRNP’s chromatin association. Six pairs of oligonucleotides were used to amplify different regions (short lines at the bottom of the top panel) of the ACT1 gene containing two exons (boxes, top panel) and an intron (connecting thin line, top panel). All data were normalized to the signal of the first oligonucleotide pair from experiments using the wild-type strain. Data represent the mean ± S.E.M. of six independent biological replicates.
Synthetic genetic array (SGA) analysis
To start the SGA screen, we first constructed a wild-type reference strain YTC1604 (MATα snr19Δ::natMX4 can1Δ::STE2pr-Sp_his5 lyp1Δ::STE3pr-LEU2 ura3Δ0 his3Δ1 leu2Δ0 met15Δ0 pRS4161_SNR19 [URA3/MET15/CEN]) as follows. Strain Y8205 (a gift from C. Boone) (64) was crossed with strain BY4741 (65) to yield a desired segregant corresponding to strain YTC1337 (MATα can1Δ::STE2pr-Sp_his5 lyp1Δ::STE3pr-LEU2 ura3Δ0 his3Δ1 leu2Δ0 met15Δ0). We then transformed pRS4161_SNR19 into strain YTC1337. Finally, to arrive at strain YTC1604, we deleted the chromosomal SNR19 by replacing it with a natMX4 selectable marker that was PCR amplified from plasmid p4339 (a gift of C. Boone). The SGA query strain YTC1605, which harbors an U1-snRNA UU mutant allele on pRS4161_UU was constructed in the same manner. With minor modifications, we followed the standard SGA procedure (64,66) by employing a high-throughput microbial array pinning robot (RoToR HDA, Singer Instruments). Strains YTC1604 and YTC1605 were independently mated with a collection of ∼4600 nonessential gene deletion strains (Standard SGA Array, a gift from C. Boone) on YPD plate overnight before selecting for diploids on YPD+G418/clonNAT twice. The resulting diploids were sporulated on plates at 22°C for 5 days. MATa haploids containing deletions of both SNR19 and the nonessential gene were selected on (SD/MSG)-His/Arg/Lys+canavanine/thialysine plates twice before image acquisition and data processing, which was done using the ScreenMill (67).
U1 snRNP purification
To purify the U1 snRNP, we first constructed strain YTC1549 (MATaSTO1–3xFLAG::hphMX6 snr19Δ::natMX4 MUD1-TAP::HIS3MX6 his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pCG106 [SNR19/URA3/CEN]). Subsequent replacement of pCG106 (50) with pSNR19001 derivatives (see above) made possible for purifying U1 snRNP variants differing at the 5′-end dinucleotide of the U1 snRNA (GG, CU, and wild-type) (Figure 6B). We followed the published protocol for tandem affinity purification of U1 snRNP via the TAP-tagged Mud1p (42), except that the calmodulin affinity step was replaced by a Mono Q step. After the TEV protease elution step from the IgG Sepharose, the eluted material was loaded onto a Mono Q 5/50 GL column (GE Healthcare Life Sciences) and the bound proteins were eluted by a linear gradient of NaCl (80–600 mM). Fractions (0.5 ml) were collected and analyzed by 8% SDS-PAGE. For Western blotting, the purified U1 snRNP was fractionated on a gradient gel (4–12%) (Invitrogen) and probed for the abundance of Sto1p using anti-FLAG M2 antibody (Sigma-Aldrich) (1:1000 dilution). The amounts of Prp40p and Smd1p in the same fractions were also determined by anti-Prp40p antibody (1: 5000 dilution) and anti-Smd1p antibody (1:1000 dilution).
Figure 6.
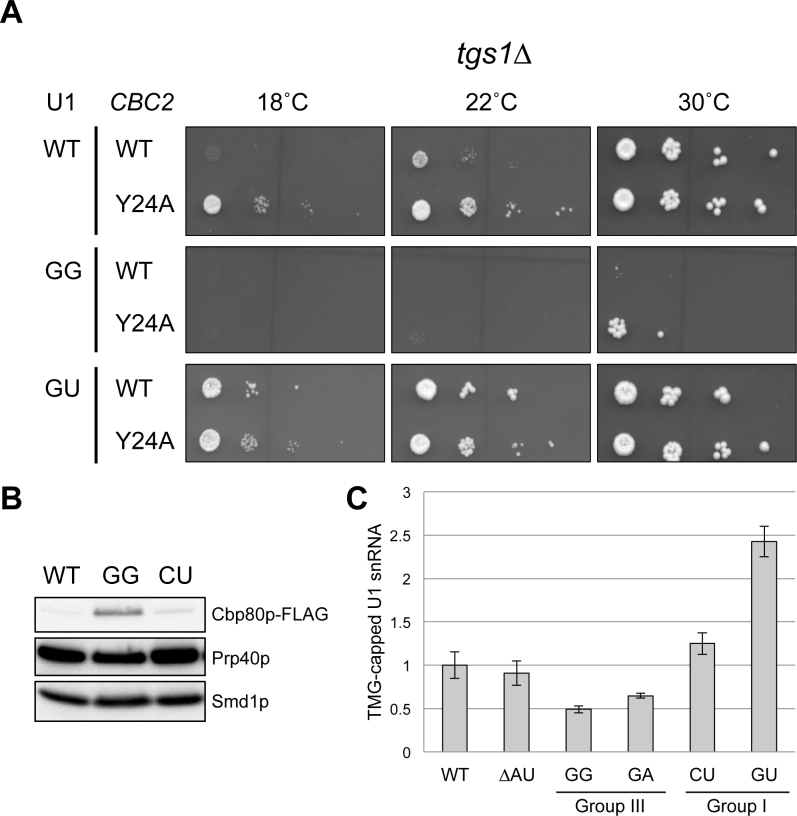
Alteration of U1 snRNA 5′-end dinucleotide impacts on CBC binding to U1 snRNA. (A) Rescue of the GG/tgs1Δ synthetic lethality by CBC2-Y24A mutation at 30°C. Yeast strains bearing tgs1Δ in conjunction with various combinations of SNR19 and CBC2 alleles were examined for their growth phenotypes at different temperatures. The GU allele that can rescue tgs1Δ cs phenotype was included as a control. (B) The U1 snRNA 5′-end GG mutant provides an environment for hyperstable interaction between its m7G cap and CBC. U1 snRNP variants differing at the 5′-end dinucleotide of the U1 snRNA (GG, CU, and wild-type) were affinity purified and probed the abundance of Cbp80p using anti-FLAG M2 antibody. Anti-Prp40p and anti-Smd1p were respectively used to detect the Prp40p and Smd1p, which are integral to U1 snRNP. (C) The identity of the U1 snRNA 5′-end dinucleotide may impact on the TMG cap formation. More U1 snRNA from the CU and GU mutants (Group I) could be precipitated by anti-TMG antibody than that of the control strains (WT and ΔAU). In contrast, less U1 snRNA could be recovered from the GG and GA mutants (Group III). The precipitated U1 snRNA was quantitated by RT-qPCR and normalized against U2 snRNA in the same RNA sample. Data represent the mean ± S.E.M. of at least three independent biological replicates.
Rescuing the GG/tgs1Δ synthetic lethality by CBC2-Y24A mutation
We first constructed strain YTC1565 (MATasnr19Δ::natMX4 tgs1Δ::kanMX4 cbc2Δ::hphMX4 can1Δ::STE2pr_Sp_his5 lyp1Δ::STE3pr_LEU2 ura3Δ0 his3Δ1 leu2Δ0 met15Δ0 pCG106), to which pCBC2001 (CBC2/MET15/CEN) and pCBC2002 (CBC2-Y24A/MET15/CEN) were individually transformed. The resulting strains were then transformed with pSNR19001 variants for expressing wild-type U1 snRNA, GG-U1 snRNA, or GU-U1 snRNA, respectively. These tester strains were initially cultured in a medium lacking leucine, methionine, and uracil to retain the all three plasmids, serially diluted, and then spotted on the 5-flouroortic acid (5-FOA) solid medium lacking leucine and methionine for counterselecting pCG106. The spotted plates were incubated at 18, 22 and 30°C until colony formation.
Quantitation of TMG-capped U1 snRNA
Yeast strains expressing various U1 snRNA variants were used for extracting total RNAs by standard hot phenol/chloroform method. An aliquot of total RNA (100–300 μg) was then incubated at 4°C for 3 h with 10 μl of Dynabeads® Protein A (Thermo Fisher), which were bound by 10 μl of anti-TMG antibody (R1131, Anti-m3G-cap, [Cat#201002], Synaptic Systems). After extensive washes, the bound RNAs were extracted and purified. The amounts of U1 and U2 snRNAs in the precipitated as well as in the input RNAs were quantitated by RT-qPCR using U1- and U2-specific primer sets, respectively. The amount of anti-TMG precipitated U1 snRNA was calculated as [U1IP/U2IP] / [U1Input/U2Input] and further normalized to the value obtained from the wild-type strain. Data from three biological repeats were averaged and shown in Figure 6C.
Molecular docking and estimation of binding free energy
In brief, the atomic structure of the human nuclear cap-binding-complex (CBC) in complex with a cap analogue m7GpppG (PDB: 1H2T) (24) was downloaded as a template. Next, the structure of CBP20 (nuclear cap-binding protein, 20 kDa) and that of m7GpppG analog (‘chain Z’ in PDB: 1H2T) were extracted respectively from the template as the ‘protein’ and ‘ligand’. To perform in silico mutation of RNA nucleotide (m7GpppG change to m7GpppA, m7GpppU, or m7GpppC) or to substitute amino acid residue (Y138A), commands of ‘swapna’ and ‘swapaa’ was used respectively in UCSF Chimera (version 10.2) (68). For further docking analysis, the atomic structures of the protein and ligand were loaded into MGLTools (version 1.5.6) (69) to create the PDBQT structure files, recording the coordinates of non-hydrogen atoms and hydrogen atoms in all polar residues, partial charges and a description of the rigid and rotatable parts of the molecule. The docking site for m7GpppG analog on a protein target was defined by a grid box that covers the entire m7GpppG analog. The binding Gibbs free energies (ΔG°) (kcal/mol) between each particular ligands and the target protein was calculated using AutoDock Vina (version 1.1.3) (70), and was compared with that extracted based on in vitro data of the cap-CBC complexes at 20°C (24,71).
RESULTS
Altering the AU dinucleotide at the 5′ end of the U1 snRNA results in a growth defect
To address the significance of the conserved AU dinucleotide at the 5′-end of the U1 snRNA (Figure 1A), 19 plasmids were constructed that enabled expression of U1 snRNA variants corresponding to all possible permutations of the 5′-terminal dinucleotides, as well as individual or pairwise deletions at these positions. These variant U1 snRNAs were then tested for their ability to support cell viability in the absence of a wild-type U1 snRNA. Unexpectedly, all 18 mutant strains were viable and appeared to grow at a rate similar to that of the wild-type strain. Quantitative measurement of their growth rates via a GFP-based competitive fitness assay (see Methods), however, revealed that many mutant strains in fact exhibited reduced fitness compared to the wild-type strain (Figure 1B). Three mutant strains in particular (GG, GA, and UU) showed severely compromised fitness (Figure 1B). For example, the growth rate of UU mutant was reduced by >40% when grown at 30°C and UU and GG mutants were cold sensitive at 16°C (Figure 1B and C).
Altering the AU dinucleotide at the 5′ end of the U1 snRNA impacts its transcription
To examine whether these plasmid-borne U1 snRNA genes were transcribed properly, we mapped the 5′ ends of the mutant U1 snRNAs by primer extension (Figure 2A). Most of the mutants produced U1 snRNAs at a level comparable to that of the wild-type strain, and their transcription start sites were either identical to that of the wild-type, shifted as expected for deletion mutants, or shifted to nearby downstream A or G residues (Figure 2A and B). However, in the UU mutant, which exhibited the lowest fitness score and displayed a cold-sensitive phenotype (Figure 1B and C), the full-length U1 snRNA was barely detectable (Figure 2A, lane 17). In this mutant, the majority of the U1 snRNAs detected appeared to lack first 19 or 29 nucleotides of the snRNA, thus entirely missing the region necessary for base-pairing with the 5′ss (Figure 1A).
To address whether the shortened U1 snRNA species in the UU mutant were packaged into U1 snRNP, we fractionated the UU mutant cell extract on a sucrose gradient and monitored U1 snRNP using an antibody targeting Prp40p, an integral U1-snRNP protein (Supplementary Figure S2). The UU mutant's U1 snRNP reproducibly peaked at fraction 8, whereas the wild-type U1 snRNP peaked at fraction 7, suggesting a conformational and/or structural change. Primer extension showed that the 5′-shortened U1 snRNA species were present in these fractions (Supplementary Figure S2), suggesting that they were packaged into Prp40p-containing U1 snRNP particles.
The fact that the UU mutant was viable, although fitness compromised, raised the possibility that the shortened U1 snRNA species might still be functional. This consideration was in light of the previous reports that addition of excess SR proteins was capable of rescuing the splicing defect upon sequestering the U1 snRNA 5′ end or depletion of U1 snRNP in vitro (72,73). To test this hypothesis and to facilitate interpretation of subsequent primer extension assays, we constructed a LEU2-marked plasmid capable of expressing an internally deleted (nucleotides 196–239) form of U1 snRNA (74) (Supplementary Figure S3A, lane 2; S3B, lane 1). Expression of this short form of U1 snRNA (U1s) in the snr19Δ cell permitted the loss of an SNR19WT/URA3 plasmid, indicating the functionality of U1s (Supplementary Figure S3C, bottom panel). On this U1s plasmid backbone, we then engineered a series of 5′-end truncations that allowed transcription of the U1s species precisely from +7, +12, +17 or +20 positions (Supplementary Figure S3B, lanes 3–6). None of these truncated U1s species were able to complement the chromosomal snr19Δ deletion (Supplementary Figure S3C). This result suggested that the 5′-end truncated U1 snRNA species were not functional and that the viability of the UU mutant was most likely derived from a sufficiently low level of full-length U1 snRNA in the cell. Quantitative analysis by primer extension of the full-length U1 snRNA abundance in all the mutants, which was normalized the intrinsic U2 snRNA levels, supported this notion (Figure 2C, see the UU lane; Supplementary Figure S2).
Altering the AU dinucleotide at the 5′ end of the U1 snRNA impacts splicing in vivo
To determine the in vivo splicing defects associated with the UU mutant we turned to a splicing-sensitive microarray platform (59,75,76). Splicing in the UU mutant was, as expected, severely compromised at 30°C, as seen by the extensive accumulation of many intron-containing transcripts in comparison to that of the wild-type strain (Figure 3A). We then used RT-qPCR to validate this genome-wide analysis and to examine whether splicing was also compromised in two other mutants, GG and GA, which also exhibited significant fitness loss (Figure 1B). The ΔAU mutant was included as a control, because it had a fitness score nearly identical to that of the wild-type strain (Figure 1B), despite producing a shortened U1 snRNA missing the first two nucleotides (Figure 2A, lane 2 and Figure 2B) (See detailed analysis and discussion of the ΔAU mutant below). A panel of nine intron-containing genes were chosen for this analysis. For each of these genes, primer pairs were used to separately amplify regions within the intron or the downstream exon, corresponding respectively to the levels of pre-mRNA (P) and total (T) transcripts. The ratio of P/T in the mutant background was then normalized against the same ratio obtained from the wild-type strain. This analysis consistently showed that pre-mRNA accumulated significantly in UU, GG, and GA mutant strains (i.e. P/T > 1), but less so in the ΔAU mutant (Figure 3B). Interestingly, in both the GG and GA mutants, the splicing defect (Figure 3B) correlated well with the fitness deficiencies (Figures 1B and 3B), yet their levels of full-length U1 snRNA were comparable in these mutants to that in wild-type strain (Figure 2A, lanes 12 and 13; Figure 2C), suggesting that the identities of the 5′-end nucleotides play a role in splicing (see below).
Figure 3.
Alteration of U1 snRNA 5′-end dinucleotide impacts on splicing in vivo. (A) Microarrays analysis of the genome-wide splicing phenotype of the UU mutant in comparison to that of the wild-type strain. T, total transcript; P, pre-mRNA; and M, spliced transcript. The two columns under each category, i.e. T, P and M, represent a pair of dye-flipped (Cy3 and Cy5) experiment of the same RNA sample, to which one dataset is inversely presented. (B) Accumulation of intron-containing transcripts in the GG, UU, and GA mutants. Splicing defect (P/T >1) of the nine selected intron-containing genes was quantitated measured by RT-qPCR. Data represent the mean ± S.E.M. of three independent biological replicates.
Altering the AU dinucleotide at the 5′ end of the U1 snRNA impacts its recruitment to pre-mRNAs
Alteration of the 5′-end dinucleotide of U1 snRNA could potentially change the physical properties and/or the functionality of U1 snRNP. To evaluate the first issue regarding physical property, we fractionated the active splicing extracts prepared from ΔAU, GG, UU, and the wild-type strains by native gel electrophoresis and performed Northern blotting using a U1-snRNA-specific probe. Two major forms of U1 snRNP were detected in all cases (Figure 4A), consistent with previous reports (16,17,19). There is no detectable difference of these two forms of U1 snRNP in terms of their gel mobilities (cf. lanes 1 and 2–4). This is in sharp contrast to the effects of eliminating Prp42p, U1A/Mud1p, or Nam8p/Mud15p, in which significant alterations of gel migration pattern or loss of the fully assembled U1 snRNP were observed (Figure 4A, lane 5) (16,17). To address the second issue regarding functionality, we examined the formation of the CCs, which represent the initial binding of the U1 snRNP to pre-mRNA and can be accumulated by leaving ATP out of the in vitro splicing reaction. Radioactively labeled pre-mRNA was incubated with splicing extracts to form CCs, which were resolved by native-gel electrophoresis. A notable increase of the CC2 mobility was observed in GG and UU reactions, but not in ΔAU reaction (Figure 4A, lanes 6–9), consistent with their splicing and fitness phenotypes in vivo. This biochemical phenotype is reminiscent of the faster-migrating CCs that are seen when U1A/Mud1p (77) or Mud2p (11) is lost, but it is opposite to the result of eliminating Cbp20p/Mud13p, which causes a decrease in CC mobility (28). We interpreted the observed faster-migrating phenotype as reflecting a change of conformation or composition of CCs in vitro.
To further probe the impact of the observed change on splicing in vivo, we carried out standard ChIP assays to measure chromatin association of the Prp40p (a proxy for the U1 snRNP) with ACT1, an endogenous intron-containing gene. Formaldehyde-cross-linked chromatin was immunoprecipitated by anti-Prp40p antibody and DNA was analyzed by RT-qPCR using six sets of primers directed to different regions of the ACT1 gene (Figure 4B). In general, U1 snRNP levels peaked close to the end of the intron and then decreased substantially in the middle of exon 2, consistent with previously reported results (19,78,79). Importantly, U1 snRNP recruitment in GG and UU mutants was significantly reduced to ∼40% of the wild-type level (Figure 4B, primer pair 3). These in vivo biochemical phenotypes were not observed in the control ΔAU mutant, consistent again with the fitness, in vivo splicing, and CC formation data. Taken together, these data suggest that alteration of the 5′-end dinucleotide of the U1 snRNA impacts on CC formation and U1 snRNP recruitment during splicing.
The 5′-end dinucleotide of the U1 snRNA is genetically linked to CC components
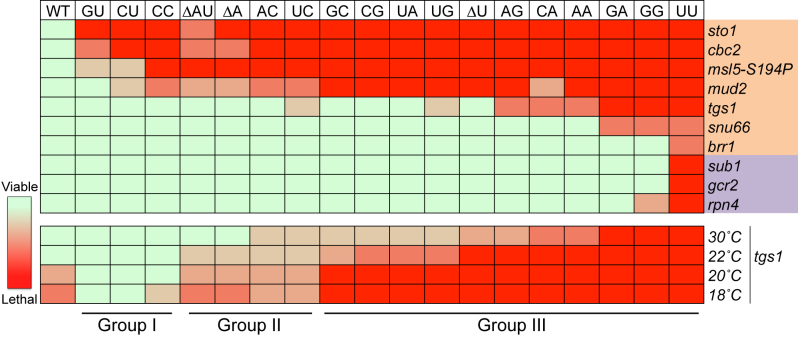
To further explore the mechanism of the splicing defect in the UU mutant (Figure 3), we performed a genome-wide synthetic-lethal screen by crossing the UU mutant with ∼4600 yeast strains, each harboring a nonessential gene deletion, and examined the growth phenotypes of the resulting double mutants through a robotic Synthetic Genetic Array (SGA) approach (66) (see Materials and Methods). This screen yielded an exceptionally high-quality dataset and led to the identification of only nine genes that are synthetically sick or lethal with the UU mutation (Figure 5, top panel, the UU column). These genes fell into two distinct functional groups: splicing (STO1, CBC2, MUD2, TGS1, SNU66 and BRR1) and transcription (SUB1, GCR2 and RPN4). This prompted us to investigate whether the remaining seventeen U1-snRNA dinucleotide mutants also showed genetic interactions with the nine candidate genes uncovered from the SGA screen. To this end, we generated all the double mutants, which correspond to each of these nine candidate gene deletion in combination with the remaining seventeen U1-snRNA mutant alleles. Strikingly, we found that practically all U1-snRNA dinucleotide mutations were either synthetically lethal or severely sick when placed in combination with sto1Δ, cbc2Δ and mud2Δ (Figure 5; rows 1, 2 and 4). Note that STO1 and CBC2 encode Cbp80p and Cbp20p respectively, the two subunits of the nuclear cap-binding complex (CBC), and are required for stable CC formation (27,32). Likewise, Mud2p is a component of CC2 and is involved in branch-site recognition with BBP (11,12). Although BBP is an essential protein and was not identified in our first-round of screening, in an independent series of experiments we further showed that specific alteration of BBP (i.e. msl5-S194P allele) also resulted in a similar pattern of synthetic lethality (Figure 5; row 3). Taken together, this dataset immediately pointed to the involvement of the 5′-end dinucleotide of the U1 snRNA in CC formation or stability, consistent with the preceding biochemical analysis (Figure 4).
Figure 5.
Genetic analysis identified genes functionally linked to the U1 snRNA 5′-end dinucleotide. Nine candidate genes (except for BBP allele msl5-S194P) marked on the right emerged first from an SGA screen using the UU mutation, among them six are related to splicing (light brown) and three to transcription (light purple). Genetic interaction of these nine candidate genes and msl5-S194P with all the U1 snRNA mutants was further manually analyzed in a pairwise manner. The severities of synthetic lethality, ranging from viable to lethal, are presented in a heat map (top half). Temperature-dependent synthetic lethality between tgs1Δ and all the U1 snRNA mutations is presented in the bottom half. See text for detailed discussion of the biological underpinning of Groups I, II and III.
A competing hypothesis would be that without functional CBC, which binds to the m7G cap of the nascent snRNAs in the nucleus, a simultaneous alteration of the 5′-end dinucleotide of the U1 snRNA may negatively impact the U1-snRNA and/or U1-snRNP biogenesis. Because the double mutants containing U1 snRNA mutants with either sto1Δ or cbc2Δ could not be analyzed due their severe growth phenotype, we exploited the aforementioned short-form U1 snRNA (U1s) system (Supplementary Figure S3A) to test this alternate possibility. We first kept the double mutants alive by introducing a plasmid expressing the full-length wild-type U1 snRNA and, under such condition, the 5′-end altered U1s snRNA species were examined by primer extension. The U1s snRNA species in the UU, AA and GG mutants were found to transcribe from the same start sites seen in the full-length context, and express at a level comparable to that of the wild-type U1s snRNA (Supplementary Figure S3A; lanes 4–11). We were, however, unable to characterize the property of the corresponding U1s snRNPs in the double mutants using the similar approach, because the presence of full-length U1 snRNP masked the potential gel-mobility changes. Regardless, our data are consistent with an interpretation that the U1 snRNA 5′-end dinucleotide plays a role in CC formation.
Our genetic analyses of the 5′-end dinucleotide mutants revealed that the UU mutation was unique in its synthetic-lethal phenotype when placed in combination with sub1Δ, gcr2Δ, or rpn4Δ (Figure 5, the UU column). This is consistent with the finding that only the UU mutant exhibited a dramatic transcriptional defect that yielded high levels of 5′ end-truncated U1 snRNAs but very low levels of full length U1 snRNA (Figure 2). We speculated that, in the UU mutant, the already limited supply of the full-length U1 snRNA (Figure 2C) may be further reduced below a threshold for supporting cell viability in the absence of either Sub1p, Gcr2p or Rpn4p. If so, these three transcription factors may have a role in regulating transcription of SNR19 or other snRNA genes.
Altering the AU dinucleotide at the 5′ end of the U1 snRNA impacts CBC binding to U1 snRNA
Another gene that emerged from our SGA screen was TGS1, which encodes trimethyl guanosine synthase, the previously described enzyme responsible for converting the m7G cap of snRNAs, snoRNAs, and telomerase RNA into a TMG cap (36,38). Interestingly, tgs1Δ is only synthetic lethal with GA, GG, and UU mutations at 30°C (Figure 5; bottom panel), which correlates well with their low fitness scores (Figure 1B). To probe the underlying mechanism for this genetic observation, we took inspiration from a recent finding that, in the absence of Tgs1p, the 5′ cap of the U1 snRNA is locked in the m7G state and, as a result, is prone to remain bound by CBC (42). It was hypothesized (42,43) that such an interaction is thermodynamically strengthened by low temperature, thus leading to the observed cold-sensitive (cs) phenotype of the tgs1Δ strain, most likely reflecting a defect associated with usage of CBC-bound U1 snRNP in the spliceosome. Subsequent experiments (42,43) using a CBC2-Y24A allele to loosen the interactions between CBC and m7G cap indeed rescued the cs phenotype. Accordingly, we wondered whether the synthetic lethality of the GG and GA mutations with tgs1Δ could also be due to an unfavorable association of CBC with the corresponding U1 snRNA variants even at the permissive temperature (30°C). If this were the case, loosening the interaction between Cbp20p and the m7G cap in the tgs1Δ background should rescue the synthetic lethality of the GG and GA mutant. To test this hypothesis, we first constructed a haploid strain bearing the following features: (1) chromosomal deletions of TGS1, CBC2, and SNR19; (2) an SNR19/URA3 plasmid; and (3) a plasmid harboring either CBC2 or CBC2-Y24A. We then transformed a plasmid-borne U1-GG or U1-GA allele into the starting strains to allow 5-FOA counterselection of the SNR19/URA3 plasmid and then subjected the resulting strains to growth phenotype assessment at various temperatures. We found that CBC2-Y24A indeed rescued tgs1Δ cs phenotype (Figure 6A; top panels) as previously reported (42,43). Remarkably, CBC2-Y24A, but not CBC2, relieved the GG/tgs1Δ and GA/tgs1Δ synthetic lethality at 30°C (Figure 6A; right panels, and Supplementary Figure S4), consistent with our prediction.
Encouraged by the preceding findings, we then surveyed all U1 snRNA 5′-end dinucleotide mutations for their potential genetic interactions with tgs1Δ at low temperatures (Figure 5; bottom panels). We found that the 5′-end dinucleotide mutations can be classified into three groups based on their behavior in the tgs1Δ context. Members in the first group (Group I), which include GU, CU and CC, were found to grow well at 18°C in the presence of tgs1Δ. A moderate degree of synthetic sickness with tgs1Δ was observed for the second group (Group II), which includes ΔAU, ΔA, AC and UC. In contrast, members in the third group (Group III), which include GC, CG, UA, UG, ΔU, AG, CA, AA, GA, GG and UU, were synthetic lethal (or severely sick) when placed in combination with tgs1Δ at 18, 20, and 22°C. Three of them, i.e., GA, GG, and UU, failed to support growth at all temperatures tested. We interpreted these genetic data as follows. First, the synthetic lethality between Group III mutations and tgs1Δ may result from a hyperstable interaction between CBC and m7G cap even at the 30°C, akin to the proposed mechanism of the cold-sensitive phenotype of tgs1Δ described above (42,43). Second, the lack of synthetic lethality between Group I mutations and tgs1Δ at 16°C would then be parallel to the observation that CBC2-Y24A (i.e., weaker interaction) is capable of alleviating the cs phenotype of tgs1Δ. We therefore hypothesized that the Group I and Group III mutations may result in, respectively, a permissive weak or a prohibitively strong interaction between their corresponding U1 snRNA variants and CBC even in the presence of Tgs1p. To directly test this hypothesis, we measured the amounts of Cbp80p associated with the affinity purified U1 snRNP under the background of TGS1 and in the presence of either CU mutation (Group I), GG mutation (Group III), or wild-type SNR19 (see Methods). Cbp80p was indeed found to be at least 3-fold higher in the U1 snRNP purified from the GG mutant than those from the CU mutant and the wild-type strain (Figure 6B).
Our working model thus predicts that the Group III snRNA variants, which are severely synthetic lethal with tgs1Δ, promote hyperstable interaction between their m7G cap and CBC, in contrast to the wild-type snRNA and the Group I snRNA variants. If this were the case, one would expect that the m7G-to-TMG conversion during U1-snRNP biogenesis would be less efficient for the Group III U1 snRNA variants, because CBC and Tgs1p compete for the same substrate, i.e. the m7G cap. We tested this hypothesis by quantitatively measuring the immunoprecipitable amount of TMG-capped U1 snRNA using anti-TMG antibody in the total RNAs of wild-type, ΔAU, Group I (CU and GU), and Group III (GG and GA) strains in the TGS1 genetic background (see Materials and Methods). Less U1 snRNA was recovered from Group III mutants (GG [49%] and GA [65%] versus wild-type value) in comparison to the controls (wild-type and ΔAU) (Figure 6C). In a striking reversal, over 1.2- and 2.4-fold more U1 snRNA was recovered from the CU and GU mutants (Group I), respectively, consistent with our hypothesis.
Taken together, this series of experiments clearly demonstrated that the identity of the U1 snRNA 5′-end dinucleotide plays a critical role in providing a platform for Tgs1p to efficiently convert the m7G cap into the TMG cap and that failing to make such a conversion results in a prolonged occupation of CBC on the m7G cap, which may in turn cause steric hindrance that reduces the ability of the mutant U1 snRNP to function in spliceosome assembly.
Altering the basepairing between the 5′-AU and the 5′ss sequence is unlikely to play a key role in fitness reduction of U1 snRNA mutants
There remains, however, a lingering question as to whether potential basepairing of this AU dinucleotide to intron sequence is critical for splicing in the native, rather than in the artificial, context. If so, altering the identity of this dinucleotide may either abolish a pre-existing or establish a new basepairing event, thereby impacting splicing in vivo. To analyze this possibility, we computationally compiled a list of potential basepairing events between mutant U1 transcripts, whose 5′-end sequences were experimentally determined (Figure 2A), and all known introns in yeast (Supplementary Table S2). We first focused on the case of the GG mutant, which exhibited the second most severe growth phenotype (Figure 1B). The U1 transcript from the GG mutant, by computational prediction, establishes new basepairing with only two introns, both of which are located in nonessential genes (Supplementary Table S2, bottom row). As a result, it seems unlikely to explain the severe growth phenotype of the GG mutant. We then looked at the other side of the coin and found that the pre-existing basepairing events within 15 essential-gene introns are to be abolished when the AU dinucleotide is altered into all possible permutations (Figure 2B). Among these 15 introns (Supplementary Table S2, first row), five were chosen for measuring their splicing efficiency in two different contexts. The first is to examine splicing in the ΔAU mutant, in which the basepairing is predicted to be abolished. The second is to alter the sequence in introns predicted to base pair with AU and then examine the splicing outcome. No significant reduction of splicing, except in the case of RPL30 (see below), was detected (Supplementary Figure S5A and SB). Surprisingly, dramatic splicing defect was observed when positions 7 and 8 in the RPL30 intron were altered (AU-to-UA) (Supplementary Figure S5B). It has been well established that the 5′ exon and the first few nucleotides of the RPL30 intron forms a structure that blocks the 5′ss and inhibits splicing (80–82). It is therefore tempting to speculate that the AU-to-UA change within the RPL30 intron might have further stabilized the known structure per se, thereby strongly inhibits splicing. Computer prediction of the secondary structure appears to be in support of this conjecture (Supplementary Figure S5C). In short, although we cannot completely rule out the possibility of the proposed mismatched and noncanonical basepairing (52–54), our assessment nonetheless suggests that altering the basepairing between the 5′-AU and the 5′ss sequence is unlikely to play a key role in reducing the fitness of the U1 snRNA mutants. Rather, the intrinsic biochemical functions of the AU dinucleotide account for most, if not all, of the fitness reduction.
DISCUSSION
In this study we sought to address a long-standing puzzle as to why the AU dinucleotide is highly conserved at the 5′ end of the U1 snRNA, in spite of a lack of an apparent role in basepairing with the 5′-ss consensus sequence. While several studies (50,51) employing artificially constructed introns have raised the possibility that the AU dinucleotide may form basepairing interaction with intron sequences and influence splicing efficiency in vitro (50,51), here we provide the first experimental demonstration that this AU dinucleotide in the native context does bear significant consequence with respect to retention of CBC on the U1 snRNP, which in turn influences the methylation state of the U1 snRNA and the utilization of the U1 snRNP.
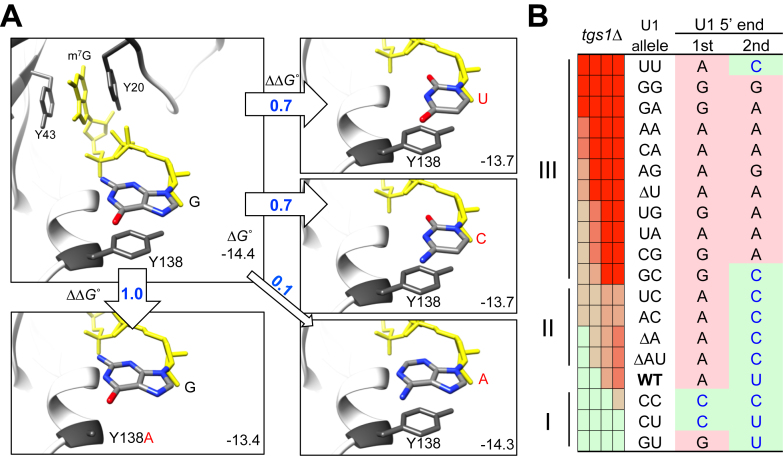
A critical observation from this study is that the identity of the 5′-end AU, when placed in conjunction with tgs1Δ, gave rise to a specific growth pattern (Figure 5). Detailed biochemical exploration (Figure 6) showed that this genetic linkage can be explained by a competition between Tgs1p and CBC for the m7G cap of the U1 snRNA, thereby influencing the methylation status of the cap structure (Figure 6). Previous structural analysis of human CBC with the cap analog m7GpppG revealed that the m7G moiety is stacked with two tyrosine residues, Y43 and Y20, and the following guanine base also stacks with Y138 (24,25,71) (Figure 7A). Experimentally altering Y138 into alanine (Y138A) was found to exact a cost of Gibbs free energy (ΔGo) by 0.8 kcal/mol and reduce the equilibrium association constant (Kas) by 1.65-fold (71) (Supplementary Table S3). Intrigued by these data, we computationally altered Y138′s stacking partner from G to either A or pyrimidine (either C or U). Changing G to A yielded a relatively minor ΔGo cost (0.1 kcal/mol), whereas changing G to either C or U led to a more pronounced expenditure (0.7 kcal/mol) comparable to that of Y138A mutation. Because Y138 is situated in a flexible loop (24,25,71), it is tempting to speculate that it may also interact with the second base (e.g., the N within m7GpppGpN), which is unfortunately not available in the structure. Should that be the case, a logical prediction of the order of CBC-binding affinity would be that of m7Gppp(G/A)p(G/A) > m7Gppp(G/A)p(C/U) > m7Gppp(C/U)p(C/U), which appears to be strikingly consistent with the observed growth pattern from our genetic analysis (Figure 7B). First, the growth defects of the Group III mutants (yielding m7GpppGpG, m7GpppGpA, m7GpppApG, or m7GpppApA end) are far more severe than those of the Group II (yielding m7GpppGpC or m7GpppApC end). Second, the growth phenotypes of the Group II mutants are comparable to that of the wild-type strain, which produces the m7GpppApU end. Third, among the three Group I mutants, CC and CU, which produce m7GpppCpC and m7GpppCpU ends, grow even better than the wild-type strain. The sole inconsistency is the GU mutant, which yields an m7GpppGpU end and is therefore predicted to be a member of the Group II. We note that the severe growth phenotype of the UU mutant is due to a substantially suboptimal level of full-length U1 snRNA in the cell (Figure 2C, see the UU lane; Supplementary Figure S2). Taken together, we propose that the conservation of the AU dinucleotide at the 5′ end of U1 snRNA is evolutionarily optimized for a need to sequentially interact with CBC and Tgs1p. In this view, the first identity (i.e. A) is selected for conforming to a purine-nucleotide start by pol II (83,84) and the second identity (i.e. U) is selected for an optimal ΔGo in favor of an efficient exchange of CBC and Tgs1p. In addition, the AU dinucleotide may also be selected for promoting optimal formation of CC during splicing (see below). Validation of this hypothesis would require determining the structure of CBC in complex with m7GpppGpN and/or m7GpppApN or performing binding assay with U1 transcript starting with m7GpppApU and alike. The latter is so far unfortunately insurmountable because transcription by T7 RNA polymerase cannot start with A, nor is it agreeable with a U at the second position (85,86).
Figure 7.
In silico analysis of stacking between m7GpppG and Y138 residue in CBP20. (A) The G base immediately following the m7Gppp moiety (shown in yellow) was replaced by either U, C or A via molecular docking and the corresponding ΔGo values were computed (see Materials and Methods) and are shown at the lower right corner within each rectangular boxes. The resulting ΔΔGo values in reference to the wild-type state are indicated within or by the empty arrows. The previously experimentally tested Y138A mutation in relationship to m7GpppG is included as a control (box, lower left). (B) Striking correlation of the genetic data with the computationally predicted ΔΔGo (see text for detailed description). The precise 5′ ends of U1 transcripts (U1 5′ end) produced in all U1 snRNA mutants (i.e. U1 allele) are shown for their respective first (1st) and second (2nd) nucleotides in each row (see also Figure 2). Note that the (C/U)-(C/U), (G/A)-(C/U), and (G/A)-(G/A) combinations are predicted to cause weaker, wild-type-like, and excessive binding between U1 snRNA and CBP20, respectively. This pattern parallels to the Group I, II, and III assignments of the tgs1Δ synthetic lethality pattern (see also Figure 5).
A prominent outcome of altering the first two nucleotides of SNR19 at the DNA level is a shift of the transcription start site (TSS) in many cases (Figure 2A and B). This is in sharp contrast to the steady-state levels of the U1 transcripts, which remain largely unchanged (Figure 2C). Essentially all these TSS shifts, however, with the sole exception of the more dramatic shifts found in the UU mutant (Supplementary Figure S3), still preserve the requirements for a canonical interaction between U1 snRNA and the 5′ss. Thus, these TSS shifts are unlikely to principally account for the observed fitness reduction. In contrast, in the specific case of the UU mutant, the majority of U1 transcripts produced are shortened to the extent that the region for forming canonical basepairing with 5′ss is lost and are therefore predicted to be nonfunctional, which is indeed the case (Supplementary Figure S3C). This finding suggests that the severe growth phenotype of the UU mutant is most likely due to the transcriptional defect that adversely reduces the steady-state level of full-length U1 snRNA. That is, the primary defect in the UU mutant is most likely at the transcriptional level. This conclusion is further supported by the finding that, in the SGA screen using the UU mutant, three genes encoding for known transcription factors were uncovered (Figure 5), which was not the case for other mutants. Notably, among the three transcription factors, Sub1p was recently reported to regulate TSS selection (87). It has been a mystery as to why and how transcription of snRNA genes by pol II tends to start at a single site with exquisite precision, whereas multiple transcriptional start sites are typically found for other pol II-transcribed genes (88). Perhaps the three transcription factors uncovered from the UU mutant screen may have a role in governing the precise start-site selection for the pol II-transcribed snRNA genes. Future research aiming to test this hypothesis should help to clarify this issue.
When considering the sequences of the U1 snRNAs from sixteen divergent organisms (Figure 1A), the sequence in S. pombe is noticeably peculiar in that the conserved AU dinucleotide is completely missing (55) and extending the S. pombe U1 snRNA with the AU dinucleotide at its 5′ end does not appear to affect cell growth (89). These observations therefore seemingly argue against the evolutionary significance of the AU dinucleotide. Yet, our data showed that a complete loss of the AU dinucleotide of the U1 snRNA transcript (ΔAU and ΔA, Figure 2B) results in a moderate yet significant growth defect in synthetic media (Supplementary Figure S6) and synthetic lethality with an msl5 mutant allele (Figure 5, row 3), supporting that the dinucleotide is critical for maintaining cellular fitness. To explain this apparent conundrum, we aligned the U1 snRNA sequences found in various species of Schizosaccharomyces (Supplementary Figure S1). Remarkably, the 5′ ends of these U1 snRNA transcripts either are missing the AU dinucleotide or have the AU dinucleotide replaced by either UU or UC. It is known that pol II strongly prefers to start transcription from a purine (83,84), usually an A, so it is likely that the reported 5′ ends in the databases, which have yet to be experimentally validated, might have been incorrectly assigned. If indeed so, it is highly plausible that the Schizosaccharomyces U1 snRNA might have undertaken a distinct evolutionary route, which results in the absence of the 5′-end AU dinucleotide.
In this study, we showed that, in the budding yeast, the 5′-end AU dinucleotide of U1 snRNA does not appear to participate in basepairing with the 5′ss region (Supplementary Figure S5 and Supplementary Table S2), consistent with a lack of sequence conservation of positions 7–8 at the 5′ss region (44,45). Intriguingly, experimentally introducing extra base pairs between the AU dinucleotide and the 5′ss impairs splicing in yeast (50), but improves splicing in humans (51). This latter finding thus suggests that the AU dinucleotide may contribute to the binding of U1 snRNP to the 5′ss in humans. Subsequent studies indeed showed that the AU dinucleotide may engage in a variety of noncanonical basepairing with the 5′ss region in vivo (53,54). Such a finding may be rationalized by the fact that the conserved cis-information at the 5′ss region in humans is restricted to GU, thus necessitates using the 5′-end AU dinucleotide to maximize the strength of the RNA duplex formed between U1 snRNA and the 5′ss to improve splicing-site utility. Alternatively, the difference may simply reflect an evolutionary adaptation to the optimal growth conditions, yeast at 30°C and humans at 37°C. In this view, hyperstabilization by extra basepairs using the AU dinucleotide, may disallows efficient departure of U1 snRNP from the 5′ss for yeast at 30°C. Consistent with this thermodynamic hypothesis, the splicing defect caused by hyperstabilization can be relieved by temperature shift in both yeast and human (50,51).
Finally, our data showed that the 5′-AU dinucleotide of the U1 snRNA is critical to the formation of CCs (Figures 4 and 5). This finding is consistent with the evidence for the existence of a complicated web of genetic and physical interactions among many intrinsic components of CCs, which include CBC, Mud2p and BBP, as well as the subsequently joined U2 snRNP (22), which leads to the formation of pre-spliceosome. We note that CBC is not only involved in U1 snRNP maturation as discussed, but also in CC formation (Figure 5) (27–30). While the detailed mechanism regarding how CBC and the 5′-AU dinucleotide may jointly influence CC formation remains unknown, recent advances in solving several spliceosome structures at different stages of the splicing pathway (90–92) hold great promise in this regard. In this light, structural resolution of early splicing complexes is expected to reveal the mechanistic underpinning on the functional interactions of the 5′-AU dinucleotide with CBC and Mud2p.
Supplementary Material
ACKNOWLEDGEMENTS
We thank C. Boone for providing the nonessential gene deletion yeast collection, strain Y8205, and plasmid p4339; J.-Y. Leu for GFP reference strain; H.-D. Chen for FLAG-tag plasmids; P. G. Siliciano for anti-Prp40p antibody and S.-C. Cheng for anti-Smd1p antibody.
Footnotes
Present addresses:
Jui-Hui Chen, Department of Biotechnology, Green Technology Research Institute, Taiwan CPC corporation, Kaohsiung, Taiwan.
Yue-Chang Chou, Institute of Biologics, Development Center for Biotechnology, Taipei, Taiwan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology [105-2627-M-001-002, 105-2311-B-001-059]; Academia Sinica Thematic Projects [AS-103-TP-B12]; Genomics Research Center, Academia Sinica (to T.-H.C.); Academia Sinica Thematic Project [AS-103-TP-B12 to W.-h.C.]; Academia Sinica Nano Program, and MOST [103-2113-M-001-014-MY3]; National Institutes of Health [GM098634 to J.A.P.]; Academia Sinica Postdoctoral Fellowship (to S.-L.C., J.-H.C.). Funding for open access charge: Ministry of Science and Technology, Taiwan.
Conflict of interest statement. None declared.
REFERENCES
- 1. Matera A.G., Wang Z.. A day in the life of the spliceosome. Nat. Rev. Mol. Cell. Biol. 2014; 15:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Will C.L., Lührmann R.. Spliceosome Structure and Function. Cold Spring Harbor Perspect. Biol. 2011; 3:a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fabrizio P., Dannenberg J., Dube P., Kastner B., Stark H., Urlaub H., Luhrmann R.. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell. 2009; 36:593–608. [DOI] [PubMed] [Google Scholar]
- 4. Will C.L., Luhrmann R.. Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol. 1997; 9:320–328. [DOI] [PubMed] [Google Scholar]
- 5. Chang T.H., Tung L., Yeh F.L., Chen J.H., Chang S.L.. Functions of the DExD/H-box proteins in nuclear pre-mRNA splicing. Biochim. Biophys. Acta. 2013; 1829:764–774. [DOI] [PubMed] [Google Scholar]
- 6. Legrain P., Seraphin B., Rosbash M.. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol. Cell. Biol. 1988; 8:3755–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Seraphin B., Rosbash M.. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989; 59:349–358. [DOI] [PubMed] [Google Scholar]
- 8. Yu Y., Maroney P.A., Denker J.A., Zhang X.H., Dybkov O., Luhrmann R., Jankowsky E., Chasin L.A., Nilsen T.W.. Dynamic regulation of alternative splicing by silencers that modulate 5′ splice site competition. Cell. 2008; 135:1224–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Michaud S., Reed R.. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 1991; 5:2534–2546. [DOI] [PubMed] [Google Scholar]
- 10. Michaud S., Reed R.. A functional association between the 5′ and 3′ splice site is established in the earliest prespliceosome complex (E) in mammals. Genes Dev. 1993; 7:1008–1020. [DOI] [PubMed] [Google Scholar]
- 11. Abovich N., Liao X.C., Rosbash M.. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 1994; 8:843–854. [DOI] [PubMed] [Google Scholar]
- 12. Abovich N., Rosbash M.. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997; 89:403–412. [DOI] [PubMed] [Google Scholar]
- 13. Arning S., Gruter P., Bilbe G., Kramer A.. Mammalian splicing factor SF1 is encoded by variant cDNAs and binds to RNA. RNA. 1996; 2:794–810. [PMC free article] [PubMed] [Google Scholar]
- 14. Zamore P.D., Patton J.G., Green M.R.. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992; 355:609–614. [DOI] [PubMed] [Google Scholar]
- 15. Lockhart S.R., Rymond B.C.. Commitment of yeast pre-mRNA to the splicing pathway requires a novel U1 small nuclear ribonucleoprotein polypeptide, Prp39p. Mol. Cell. Biol. 1994; 14:3623–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McLean M.R., Rymond B.C.. Yeast pre-mRNA splicing requires a pair of U1 snRNP-associated tetratricopeptide repeat proteins. Mol. Cell. Biol. 1998; 18:353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gottschalk A., Tang J., Puig O., Salgado J., Neubauer G., Colot H.V., Mann M., Seraphin B., Rosbash M., Luhrmann R. et al. . A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA. 1998; 4:374–393. [PMC free article] [PubMed] [Google Scholar]
- 18. Du H., Rosbash M.. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature. 2002; 419:86–90. [DOI] [PubMed] [Google Scholar]
- 19. Hage R., Tung L., Du H., Stands L., Rosbash M., Chang T.H.. A targeted bypass screen identifies Ynl187p, Prp42p, Snu71p, and Cbp80p for stable U1 snRNP/Pre-mRNA interaction. Mol. Cell. Biol. 2009; 29:3941–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schwer B., Chang J., Shuman S.. Structure-function analysis of the 5′ end of yeast U1 snRNA highlights genetic interactions with the Msl5*Mud2 branchpoint-binding complex and other spliceosome assembly factors. Nucleic Acids Res. 2013; 41:7485–7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schwer B., Shuman S.. Structure-function analysis of the Yhc1 subunit of yeast U1 snRNP and genetic interactions of Yhc1 with Mud2, Nam8, Mud1, Tgs1, U1 snRNA, SmD3 and Prp28. Nucleic Acids Res. 2014; 42:4697–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schwer B., Shuman S.. Structure-function analysis and genetic interactions of the Yhc1, SmD3, SmB, and Snp1 subunits of yeast U1 snRNP and genetic interactions of SmD3 with U2 snRNP subunit Lea1. RNA. 2015; 21:1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gonatopoulos-Pournatzis T., Cowling V.H.. Cap-binding complex (CBC). Biochem. J. 2014; 457:231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mazza C., Segref A., Mattaj I.W., Cusack S.. Large‐scale induced fit recognition of an m7GpppG cap analogue by the human nuclear cap‐binding complex. EMBO J. 2002; 21:5548–5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Calero G., Wilson K.F., Ly T., Rios-Steiner J.L., Clardy J.C., Cerione R.A.. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat. Struct. Biol. 2002; 9:912–917. [DOI] [PubMed] [Google Scholar]
- 26. Mazza C., Ohno M., Segref A., Mattaj I.W., Cusack S.. Crystal structure of the human nuclear cap binding complex. Mol. Cell. 2001; 8:383–396. [DOI] [PubMed] [Google Scholar]
- 27. Lewis J.D., Görlich D., Mattaj I.W.. A yeast cap binding protein complex (yCBC) acts at an early step in pre-mRNA splicing. Nucleic Acids Res. 1996; 24:3332–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Colot H.V., Stutz F., Rosbash M.. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev. 1996; 10:1699–1708. [DOI] [PubMed] [Google Scholar]
- 29. Lewis J.D., Izaurralde E., Jarmolowski A., McGuigan C., Mattaj I.W.. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 1996; 10:1683–1698. [DOI] [PubMed] [Google Scholar]
- 30. Izaurralde E., Lewis J., McGuigan C., Jankowska M., Darzynkiewicz E., Mattaj I.W.. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994; 78:657–668. [DOI] [PubMed] [Google Scholar]
- 31. Qiu Z.R., Chico L., Chang J., Shuman S., Schwer B.. Genetic interactions of hypomorphic mutations in the m7G cap-binding pocket of yeast nuclear cap binding complex: an essential role for Cbc2 in meiosis via splicing of MER3 pre-mRNA. RNA. 2012; 18:1996–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fortes P., Kufel J., Fornerod M., Polycarpou-Schwarz M., Lafontaine D., Tollervey D., Mattaj I.W.. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol. Cell Biol. 1999; 19:6543–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fortes P., Bilbao-Cortes D., Fornerod M., Rigaut G., Raymond W., Seraphin B., Mattaj I.W.. Luc7p, a novel yeast U1 snRNP protein with a role in 5′ splice site recognition. Genes Dev. 1999; 13:2425–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Izaurralde E., Stepinski J., Darzynkiewicz E., Mattaj I.W.. A cap binding protein that may mediate nuclear export of RNA polymerase II-transcribed RNAs. J. Cell Biol. 1992; 118:1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Will C.L., Luhrmann R.. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001; 13:290–301. [DOI] [PubMed] [Google Scholar]
- 36. Hausmann S., Zheng S., Costanzo M., Brost R.L., Garcin D., Boone C., Shuman S., Schwer B.. Genetic and biochemical analysis of yeast and human cap trimethylguanosine synthase: functional overlap of 2,2,7-trimethylguanosine caps, small nuclear ribonucleoprotein components, pre-mRNA splicing factors, and RNA decay pathways. J. Biol. Chem. 2008; 283:31706–31718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mouaikel J., Bujnicki J.M., Tazi J., Bordonne R.. Sequence-structure-function relationships of Tgs1, the yeast snRNA/snoRNA cap hypermethylase. Nucleic Acids Res. 2003; 31:4899–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mouaikel J., Verheggen C., Bertrand E., Tazi J., Bordonne R.. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell. 2002; 9:891–901. [DOI] [PubMed] [Google Scholar]
- 39. Kuersten S., Ohno M., Mattaj I.W.. Nucleocytoplasmic transport: ran, beta and beyond. Trends Cell Biol. 2001; 11:497–503. [DOI] [PubMed] [Google Scholar]
- 40. Komeili A., O'Shea E.K.. New perspectives on nuclear transport. Annu. Rev. Genet. 2001; 35:341–364. [DOI] [PubMed] [Google Scholar]
- 41. Ohno M., Kataoka N., Shimura Y.. A nuclear cap binding protein from HeLa cells. Nucleic Acids Res. 1990; 18:6989–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schwer B., Erdjument-Bromage H., Shuman S.. Composition of yeast snRNPs and snoRNPs in the absence of trimethylguanosine caps reveals nuclear cap binding protein as a gained U1 component implicated in the cold-sensitivity of tgs1Delta cells. Nucleic Acids Res. 2011; 39:6715–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Qiu Z.R., Schwer B., Shuman S.. Two routes to genetic suppression of RNA trimethylguanosine cap deficiency via C-terminal truncation of U1 snRNP subunit Snp1 or overexpression of RNA polymerase subunit Rpo26. G3 (Bethesda). 2015; 5:1361–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carmel I., Tal S., Vig I., Ast G.. Comparative analysis detects dependencies among the 5′ splice-site positions. RNA. 2004; 10:828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ast G. How did alternative splicing evolve. Nat. Rev. Genet. 2004; 5:773–782. [DOI] [PubMed] [Google Scholar]
- 46. Kondo Y., Oubridge C., van Roon A.M., Nagai K.. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. Elife. 2015; 4, doi:10.7554/eLife.04986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Siliciano P.G., Guthrie C.. 5′ splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988; 2:1258–1267. [DOI] [PubMed] [Google Scholar]
- 48. Seraphin B., Kretzner L., Rosbash M.. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J. 1988; 7:2533–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhuang Y., Weiner A.M.. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986; 46:827–835. [DOI] [PubMed] [Google Scholar]
- 50. Staley J.P., Guthrie C.. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell. 1999; 3:55–64. [DOI] [PubMed] [Google Scholar]
- 51. Freund M., Hicks M.J., Konermann C., Otte M., Hertel K.J., Schaal H.. Extended base pair complementarity between U1 snRNA and the 5′ splice site does not inhibit splicing in higher eukaryotes, but rather increases 5′ splice site recognition. Nucleic Acids Res. 2005; 33:5112–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Roca X., Krainer A.R., Eperon I.C.. Pick one, but be quick: 5′ splice sites and the problems of too many choices. Genes Dev. 2013; 27:129–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Roca X., Akerman M., Gaus H., Berdeja A., Bennett C.F., Krainer A.R.. Widespread recognition of 5′ splice sites by noncanonical base-pairing to U1 snRNA involving bulged nucleotides. Genes Dev. 2012; 26:1098–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tan J., Ho J.X.J., Zhong Z., Luo S., Chen G., Roca X.. Noncanonical registers and base pairs in human 5′ splice-site selection. Nucleic Acids Res. 2016; 44:3908–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Porter G., Brennwald P., Wise J.A.. U1 small nuclear RNA from Schizosaccharomyces pombe has unique and conserved features and is encoded by an essential single-copy gene. Mol. Cell Biol. 1990; 10:2874–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chang S.-L., Leu J.-Y.. A tradeoff drives the evolution of reduced metal resistance in natural populations of yeast. PLoS Genet. 2011; 7:e1002034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schwer B., Mao X., Shuman S.. Accelerated mRNA decay in conditional mutants of yeast mRNA capping enzyme. Nucleic Acids Res. 1998; 26:2050–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Inada M., Pleiss J.A.. Genome-wide approaches to monitor pre-mRNA splicing. Methods Enzymol. 2010; 470:51–75. [DOI] [PubMed] [Google Scholar]
- 59. Pleiss J.A., Whitworth G.B., Bergkessel M., Guthrie C.. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol. Cell. 2007; 27:928–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stevens S.W., Abelson J.. Yeast pre-mRNA splicing: methods, mechanisms, and machinery. Methods Enzymol. 2002; 351:200–220. [DOI] [PubMed] [Google Scholar]
- 61. Lin R.J., Newman A.J., Cheng S.C., Abelson J.. Yeast mRNA splicing in vitro. J. Biol. Chem. 1985; 260:14780–14792. [PubMed] [Google Scholar]
- 62. Umen J.G., Guthrie C.. A novel role for a U5 snRNP protein in 3[prime] splice site selection. Genes Dev. 1995; 9:855–868. [DOI] [PubMed] [Google Scholar]
- 63. Kretzner L., Rymond B.C., Rosbash M.. S. cerevisiae U1 RNA is large and has limited primary sequence homology to metazoan U1 snRNA. Cell. 1987; 50:593–602. [DOI] [PubMed] [Google Scholar]
- 64. Tong A.H.Y., Boone C. Ian Stansfield . Ian Stansfield. Michael J.R.S. Methods in Microbiology. 2007; 36, Academic Press; 369–707. [Google Scholar]
- 65. Baker Brachmann C., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D.. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998; 14:115–132. [DOI] [PubMed] [Google Scholar]
- 66. Tong A.H.Y., Boone C.. Methods in molecular biology. 2006; 313:2nd edn.171–191. [DOI] [PubMed] [Google Scholar]
- 67. Dittmar J.C., Reid R.J.D., Rothstein R.. ScreenMill: A freely available software suite for growth measurement, analysis and visualization of high-throughput screen data. BMC Bioinformatics. 2010; 11:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 69. Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J.. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009; 30:2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Trott O., Olson A.J.. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010; 31:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Worch R., Jankowska-Anyszka M., Niedzwiecka A., Stepinski J., Mazza C., Darzynkiewicz E., Cusack S., Stolarski R.. Diverse role of three tyrosines in binding of the RNA 5′ cap to the human nuclear cap binding complex. J. Mol. Biol. 2009; 385:618–627. [DOI] [PubMed] [Google Scholar]
- 72. Tarn W.Y., Steitz J.A.. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev. 1994; 8:2704–2717. [DOI] [PubMed] [Google Scholar]
- 73. Crispino J.D., Blencowe B.J., Sharp P.A.. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science. 1994; 265:1866–1869. [DOI] [PubMed] [Google Scholar]
- 74. Siliciano P.G., Kivens W.J., Guthrie C.. More than half of yeast U1 snRNA is dispensable for growth. Nucleic Acids Res. 1991; 19:6367–6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Burckin T., Nagel R., Mandel-Gutfreund Y., Shiue L., Clark T.A., Chong J.L., Chang T.H., Squazzo S., Hartzog G., Ares M. Jr. Exploring functional relationships between components of the gene expression machinery. Nat. Struct. Mol. Biol. 2005; 12:175–182. [DOI] [PubMed] [Google Scholar]
- 76. Pleiss J.A., Whitworth G.B., Bergkessel M., Guthrie C.. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007; 5:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Liao X.C., Tang J., Rosbash M.. An enhancer screen identifies a gene that encodes the yeast U1 snRNP A protein: implications for snRNP protein function in pre-mRNA splicing. Genes Dev. 1993; 7:419–428. [DOI] [PubMed] [Google Scholar]
- 78. Gornemann J., Kotovic K.M., Hujer K., Neugebauer K.M.. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell. 2005; 19:53–63. [DOI] [PubMed] [Google Scholar]
- 79. Lacadie S.A., Rosbash M.. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol. Cell. 2005; 19:65–75. [DOI] [PubMed] [Google Scholar]
- 80. Li H., Dalal S., Kohler J., Vilardell J., White S.A.. Characterization of the pre-mRNA binding site for yeast ribosomal protein L32: the importance of a purine-rich internal loop. J. Mol. Biol. 1995; 250:447–459. [DOI] [PubMed] [Google Scholar]
- 81. Dabeva M.D., Warner J.R.. Ribosomal protein L32 of Saccharomyces cerevisiae regulates both splicing and translation of its own transcript. J. Biol. Chem. 1993; 268:19669–19674. [PubMed] [Google Scholar]
- 82. Eng F.J., Warner J.R.. Structural basis for the regulation of splicing of a yeast messenger RNA. Cell. 1991; 65:797–804. [DOI] [PubMed] [Google Scholar]
- 83. Shultzaberger R.K., Chen Z., Lewis K.A., Schneider T.D.. Anatomy of Escherichia coli sigma70 promoters. Nucleic Acids Res. 2007; 35:771–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhang Z., Dietrich F.S.. Mapping of transcription start sites in Saccharomyces cerevisiae using 5′ SAGE. Nucleic Acids Res. 2005; 33:2838–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gößringer M., Helmecke D., Köhler K., Schön A., Kirsebom L.A., Bindereif A., Hartmann R.K.. Handbook of RNA Biochemistry. 2014; Wiley-VCH Verlag GmbH & Co KGaA; 1–28. [Google Scholar]
- 86. Milligan J.F., Uhlenbeck O.C.. [5]Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989; 180:51–62. [DOI] [PubMed] [Google Scholar]
- 87. Braberg H., Jin H., Moehle E.A., Chan Y.A., Wang S., Shales M., Benschop J.J., Morris J.H., Qiu C., Hu F. et al. . From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell. 2013; 154:775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kuehner J.N., Brow D.A.. Quantitative analysis of in vivo initiator selection by yeast RNA polymerase II supports a scanning model. J. Biol. Chem. 2006; 281:14119–14128. [DOI] [PubMed] [Google Scholar]
- 89. Alvarez C.J., Romfo C.M., Vanhoy R.W., Porter G.L., Wise J.A.. Mutational analysis of U1 function in Schizosaccharomyces pombe: pre-mRNAs differ in the extent and nature of their requirements for this snRNA in vivo. RNA. 1996; 2:404–418. [PMC free article] [PubMed] [Google Scholar]
- 90. Yan C., Wan R., Bai R., Huang G., Shi Y.. Structure of a yeast step II catalytically activated spliceosome. Science. 2017; 355:149–155. [DOI] [PubMed] [Google Scholar]
- 91. Fica S.M., Oubridge C., Galej W.P., Wilkinson M.E., Bai X.-C., Newman A.J., Nagai K.. Structure of a spliceosome remodelled for exon ligation. Nature. 2017; 542:377–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bertram K., Agafonov D.E., Liu W.-T., Dybkov O., Will C.L., Hartmuth K., Urlaub H., Kastner B., Stark H., Lührmann R.. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature. 2017; 542:318–323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.