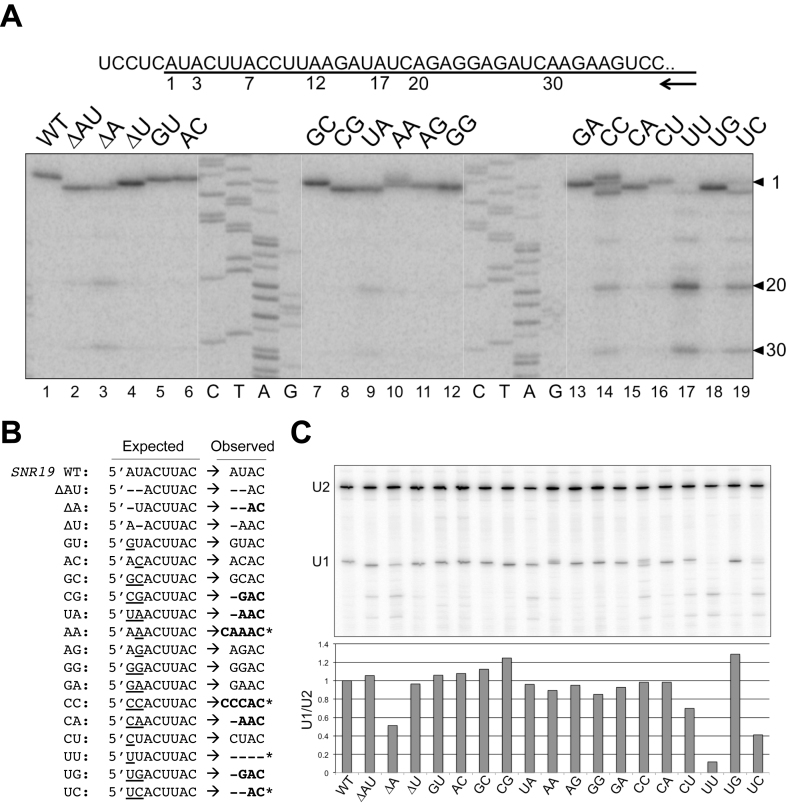
Figure 2.
Alteration of U1 snRNA 5′-end dinucleotide impacts U1 snRNA transcription. (A) Transcription start sites of the U1 snRNA variants were mapped by primer extension. Sequence around the U1 snRNA transcription start is shown in the top panel, with the U1 snRNA sequence underlined. The numbers (1, 3, 7, 12, 17, 20 and 30) below the line indicate the wild-type and variant U1 snRNA start sites determined. The arrow represents the oligonucleotide primer used in the reverse transcription reactions, which is complementary to U1 snRNA positions 266–285. Image of the primer extension results are shown in the bottom panel. The sequencing ladder (C, T, A, G) was generated by Sanger sequencing off a plasmid harboring SNR19 using the same primer. (B) Summary of the transcription start sites used by U1 snRNA variants. Deletions are marked as short dash lines and base substitutions are underlined. Variants that have unexpected start sites are shown in bold, and those which have multiple strong start sites are marked by stars. For example, AA and CC variants have a significant start from the -1 position. We noted the U1 snRNA transcription appears to prefer A and G, although there are three cases of C start (AA, CC, and CU mutants). (C) Quantitation of the full-length U1 snRNA levels in U1 snRNA mutants by primer extension normalized to the U2 snRNA levels.