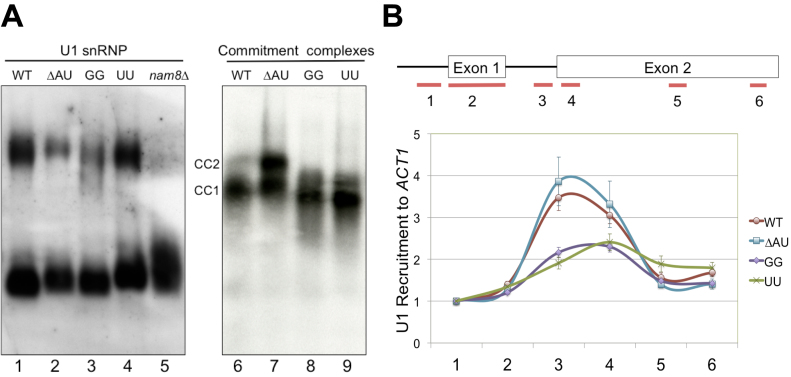
Figure 4.
Alteration of U1 snRNA 5′-end dinucleotide impacts on U1 snRNP recruitment to pre-mRNA. (A) Native gel electrophoresis analysis of U1 snRNP and commitment complexes. (Left Panel) Alteration of U1 snRNA 5′-end dinucleotide does not appear to affect U1 snRNP mobility on the native gel. Splicing extracts made from wild-type, ΔAU, UU, GG and nam8Δ strains were electrophoresed on a native polyacrylamide (3%)–agarose (0.5%) gel. The RNAs were transferred to a membrane and probed by a digoxigenin-labeled U1 probe. No apparent differences were detected for the two major forms of the U1 snRNP except for nam8Δ. (Right Panel) Electrophoretic mobilities of CCs are altered in the presence of GG and UU mutations. CC1 and CC2 are two commitment complexes formed in the absence of U2 snRNP with [32P]-labeled RP51A transcript. (B) The GG and UU mutations severely reduce U1 snRNP’s chromatin association. Six pairs of oligonucleotides were used to amplify different regions (short lines at the bottom of the top panel) of the ACT1 gene containing two exons (boxes, top panel) and an intron (connecting thin line, top panel). All data were normalized to the signal of the first oligonucleotide pair from experiments using the wild-type strain. Data represent the mean ± S.E.M. of six independent biological replicates.