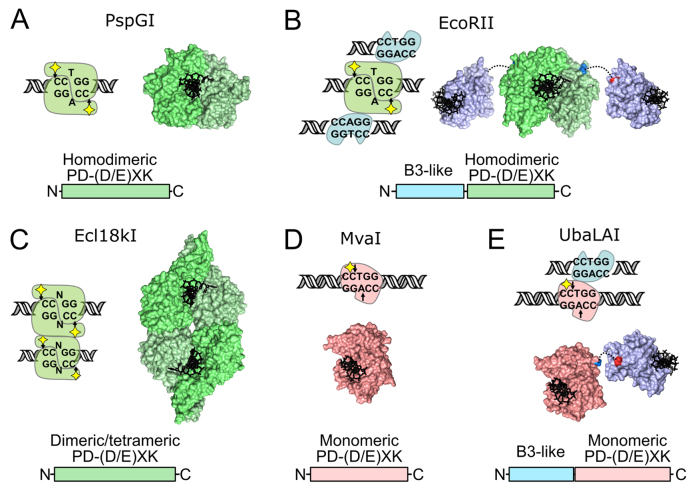
Figure 1.
Diversity of REases specific for the 5′-CCNGG-3′ and 5′-CCWGG-3′ recognition sites. In all panels catalytic PD-(D/E)XK domains in the central base pair flipping enzymes PspGI, EcoRII and Ecl18kI (36) are colored in different shades of green, monomeric MvaI-like PD-(D/E)XK domains are red, B3-like domains are blue. Yellow diamonds in cartoons denote the catalytic center. (A) PspGI (recognition sequence 5′-/CCWGG-3′) is an orthodox homodimeric REase that binds a single DNA copy (PDB ID 3bm3 (4)). (B) EcoRII (5′-/CCWGG-3′) is a homodimeric type IIE enzyme, capable of simultaneous binding of three recognition sites. One is cleaved by the PspGI-like dimer of the catalytic C-domains, while two others, one per each EcoRII-N effector domain, stimulate cleavage of the first site (PDB IDs 3hqf and 3hqg (5,7)). (C) Ecl18kI (5′-/CCNGG-3′) is a type IIF enzyme, which forms a tetramer on the DNA and simultaneously cuts both recognition sites (PDB ID 2fqz (9,10)). (D) MvaI (5′-CC/WGG-3′), like the related enzyme BcnI (5′-CC/SGG-3′), is a monomeric enzyme that uses a single catalytic center to cleave sequentially the first and then the second DNA strands (PDB ID 2oaa (11,12,14)). (E) UbaLAI (5′-CC/WGG-3′) is a monomeric REase consisting of an MvaI-like catalytic domain (red) and an EcoRII-N-like effector domain (blue, PDB ID 5o63). Structure of the UbaLAI-C domain is a model built using Modeller (37) and an MvaI-UbaLAI-C alignment generated with HHpred (38).