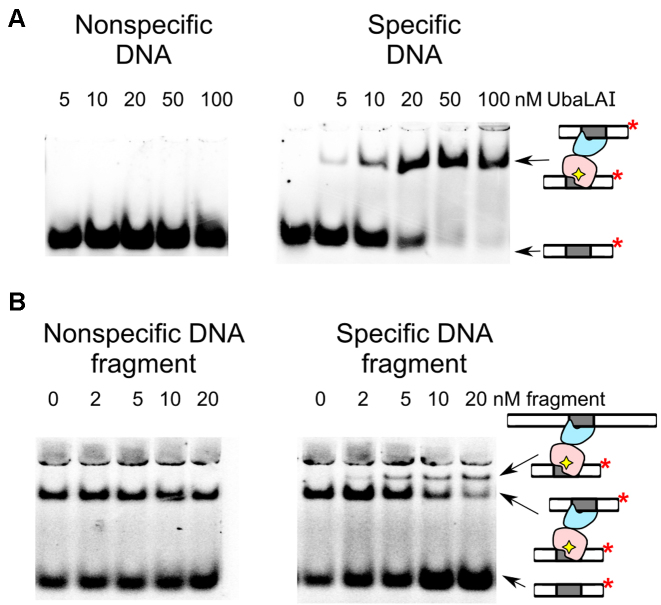
Figure 2.
UbaLAI binding analysed by electrophoretic mobility shift (EMSA) assay. (A) EMSA experiments were performed in a pH 8.3 Tris-acetate buffer supplemented with 5 mM Ca2+ (see Materials and Methods for details). The reactions contained 33P-labeled duplex (10 nM), either the non-specific 16NSP (left-hand gel) or the specific 16SP (right-hand gels), and wt UbaLAI. The protein concentrations (in terms of nM monomer) were as indicated by each lane. (B) Stoichiometry of the specific UbaLAI-DNA complex. The reactions contained 10 nM cognate 33P-labeled oligoduplex 16SP, 20 nM wt UbaLAI enzyme, and the unlabeled DNA fragment that either contains (right-hand gel) or does not contain (left-hand gel) an UbaLAI recognition site. The concentrations of the DNA fragments are noted above the relevant lanes. The cartoons on the right-hand side of the gels denote radiolabeled DNA and various types of radiolabeled complexes detected in the experiments.