Abstract
Faithful expression of transgenes in cell cultures and mice is often challenged by locus dependent epigenetic silencing. We investigated silencing of Tet-controlled expression cassettes within the mouse ROSA26 locus. We observed pronounced DNA methylation of the Tet promoter concomitant with loss of expression in mES cells as well as in differentiated cells and transgenic animals. Strikingly, the ROSA26 promoter remains active and methylation free indicating that this silencing mechanism specifically affects the transgene, but does not spread to the host's chromosomal neighborhood. To reactivate Tet cassettes a synthetic fusion protein was constructed and expressed in silenced cells. This protein includes the enzymatic domains of ten eleven translocation methylcytosine dioxygenase 1 (TET-1) as well as the Tet repressor DNA binding domain. Expression of the synthetic fusion protein and Doxycycline treatment allowed targeted demethylation of the Tet promoter in the ROSA26 locus and in another genomic site, rescuing transgene expression in cells and transgenic mice. Thus, inducible, reversible and site-specific epigenetic modulation is a promising strategy for reactivation of silenced transgene expression, independent of the integration site.
INTRODUCTION
Synthetic gene regulation systems have been designed to accurately control onset and level of transgene expression. They have been successfully employed to investigate the functions of genes or to express toxic gene products. Within the scope of synthetic biology they are of particular interest since they can be connected to endogenous regulatory networks to induce different cell fates and to control complex biological features such as proliferation (1), differentiation (2), reprogramming (3) or transdifferentiation (4,5) (reviewed in (6,7)).
Transcriptional control systems are of particular relevance since they are modular and realize inducibility of coding and regulatory RNAs. They consist of a synthetic promoter which is activated upon specific binding of activator proteins. Binding to the DNA depends on the presence of a specific trigger, which could be a small molecule, or physical cues such as electric potential or light (reviewed in (8)). One of the best characterized systems is the Tet system (9). The most frequently used synthetic Tet promoter consists of a minimal CMV promoter fused to repeats of the Tet-operator sequence from Escherichia coli. This promoter is activated upon Tetracycline or Doxycycline dependent binding of a coexpressed synthetic transactivator (rtTA), a fusion protein carrying the DNA binding domain of the tetracycline repressor protein and the activation domain of the HSV VP16 protein (10). The Tet system has also been successfully implemented in transgenic mice (11). Applications include the reprogramming of somatic cells (12), guiding cellular differentiation (13,14), overexpression of GPCR (15), Cas9 (16) or shRNA (17–19) and the labeling of cells (20).
However, despite the benefit of synthetic regulatory systems in various studies their application is hampered by unforeseen expression properties. This often concerns the loss of expression during cultivation or differentiation of transduced cells, which is thought to be the consequence of epigenetic modulation of the synthetic cassette. Since silencing of identical transgene cassettes in different integration sites differs, there seems to be a specific crosstalk between the transgene and its chromosomal surroundings.
Gene silencing is mediated by specific epigenetic modifications. DNA methylation as well as histone modifications are epigenetic marks known to influence gene expression (21). Methylation of CpG residues is mediated by DNA methyl transferases (DNMTs) and is associated with transcriptional silencing (22). Active demethylation of DNA depends on ten eleven translocation proteins (TET1–3) (reviewed in (23)). The TET protein family converts methylated cytosines (5mC) to hydroxymethylated cytosines (5hmC), a crucial step for initiating DNA demethylation (24) (reviewed in (25–27)). Next to DNA methylation, various reversible histone modifications have been described to contribute to the temporal modulation of the transcriptional activity of genes (21). These include methylation, acetylation and phosphorylation of specific histone chains, establishing the so-called histone code (28). While DNA methylation is considered to be associated with stable long-term repression, histone modifications seem to be more dynamic (21).
Synthetic Tet cassettes have been reported to be particularly susceptible for epigenetic silencing limiting applicability of this potent system (29–31). To study and overcome silencing of Tet dependent expression, we employed a defined system that allowed targeted integration of Tet cassettes into the ROSA26 chromosomal site of murine ES cells. While this locus has been shown to be ubiquitously expressed providing an open chromatin structure (32,33), the targeted Tet cassettes displayed a heterogeneous, mosaic type of expression reflected by inducible and non-inducible cells. Inducibility of the Tet cassettes was successively lost upon in vitro cultivation and differentiation, in particular in transgenic mice. We show that silencing is associated with pronounced DNA methylation of the Tet promoter. By targeting the catalytic domain of TET-1 dioxygenase to the Tet promoter we could induce targeted demethylation and re-activation of expression in murine ES cells, differentiated cells and transgenic mice.
MATERIALS AND METHODS
Cells and culture conditions
G4B12 murine embryonic stems cells (34) and targeted cell populations derived thereof were cultured in the standard cell culture medium used for murine embryonic stem cells based on DMEM (GIBCO 61965) basal medium supplemented with 15% heat-inactivated FBS (SIGMA F7524), 1× sodium pyruvate (Gibco 11360), 1x non-essential amino acids (Gibco 11140), penicillin (10 U/ml), streptomycin sulfate (100 μg/ml), 0.1M ß-mercaptoethanol (GIBCO 31350), and leukemia inhibitory factor (LIF) (1000U/ml). Selection for targeted integration was performed with 0.4 mg/ml G418. All ES cell cultures were maintained on fibroblast feeders at 37°C in a humidified atmosphere with 5% CO2 and 2% O2.
For differentiation the ES cells were seeded on a gelatinized 12-well plates (0.1% gelatin, Sigma G1393) and cultivated in DMEM supplemented with 15% fetal calf serum, 2 mM l-glutamine, penicillin (10 U/ml) and streptomycin sulfate (100 μg/ml) but without LIF at 37°C in a humidified atmosphere with 5% CO2 and 20% O2.
Rosa-Luc ES cells were created from G4B12 cells by lentiviral transduction of Cre and mCherry using pLM-CMV-R-cre (35) (gift from Michel Sadelain, Addgene P#27546). Transduced cells were sorted for mCherry and analysed for luciferase expression. Since the luciferase gene in G4B12 cells is flanked by two inverted loxP sites, in these cells the luciferase gene is permanently turned.
Mouse embryonic fibroblasts were isolated from day 13.5 embryos and immortalized upon lentiviral transfer of SV40 T antigen according to a previously published protocol (1).
For generation of BidiTet-Luc/GFP+TET1c-rtTA cells BidiTet-Luc/GFP cells were infected with a lentivirus carrying Tet1c-rttA/dsRed construct. The infected fibroblasts were sorted for dsRED expression and cultivated at 37°C in a humidified atmosphere with 7% CO2. Culture media were supplemented with 15% fetal calf serum, penicillin (10 U/ml), streptomycin sulfate (100 μg/ml), 0.1M ß-mercaptoethanol (GIBCO 31350), 1× non-essential amino acids (Gibco 11140), 1× sodium pyruvate (Sigma), 10 mM HEPES.
Doxycycline was dissolved in ethanol and administered to the cells at a final concentration of 2 μg/ml for 2 days prior to analysis. For the other chemical treatments 1 × 104 or 1 × 105 cells were seeded in a 24- or 12-well plate format, respectively. DNMT inhibitors Azacytidine (Sigma Aldrich) and Decitabine (Sigma Aldrich) were dissolved in PBS and added to the ES cells at a final molecular concentration of 1 or 0.5 μM, respectively. For the fibroblasts treatment with 1 μM Decitabine was used. Note that the incubation with epigenetic modifiers was not well tolerated by the cells which impaired extended incubation times or higher concentrations (Supplementary Figure S1A–C). Vitamin C was dissolved in PBS and used at a final concentration of 100 μg/ml. For all treatments, cells were harvested after 72 h of cultivation in the respective conditions.
Plasmids
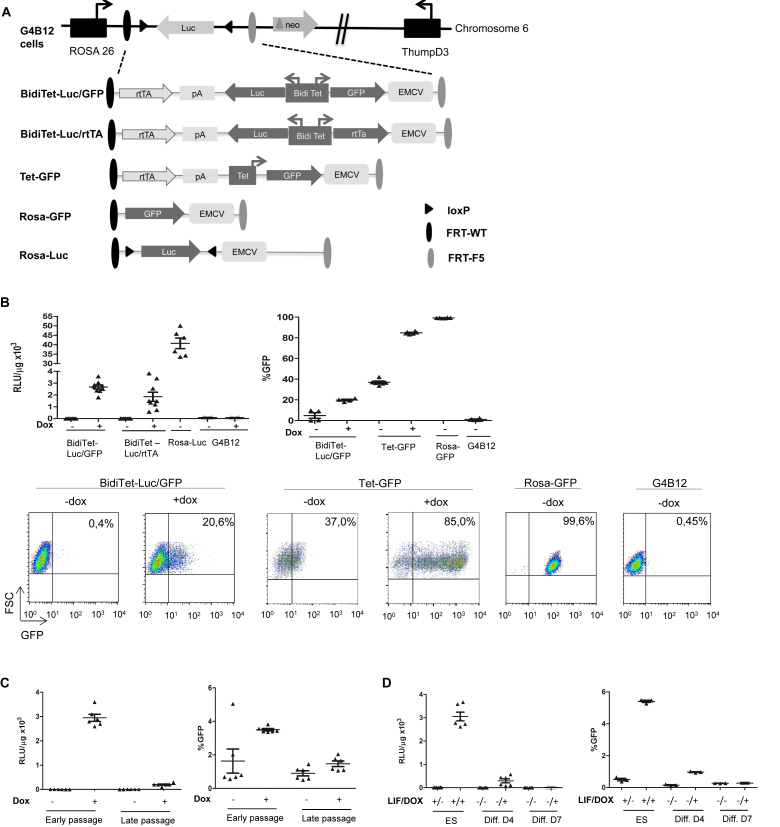
The targeting vectors pBidiTet-Luc/GFP, pBidiTet-Luc/rtTA, pRosaGFP and pTetGFP are based on pEMTAR (36) and harbor an encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) element as well as a ATG translation initiation codon. Upon targeting the cassettes in G4B12 cells the ATG is positioned in frame to the neomycin phosphotransferase gene, thereby rendering the cells resistant towards G418 (cf. Figure 1 and (37)).
Figure 1.
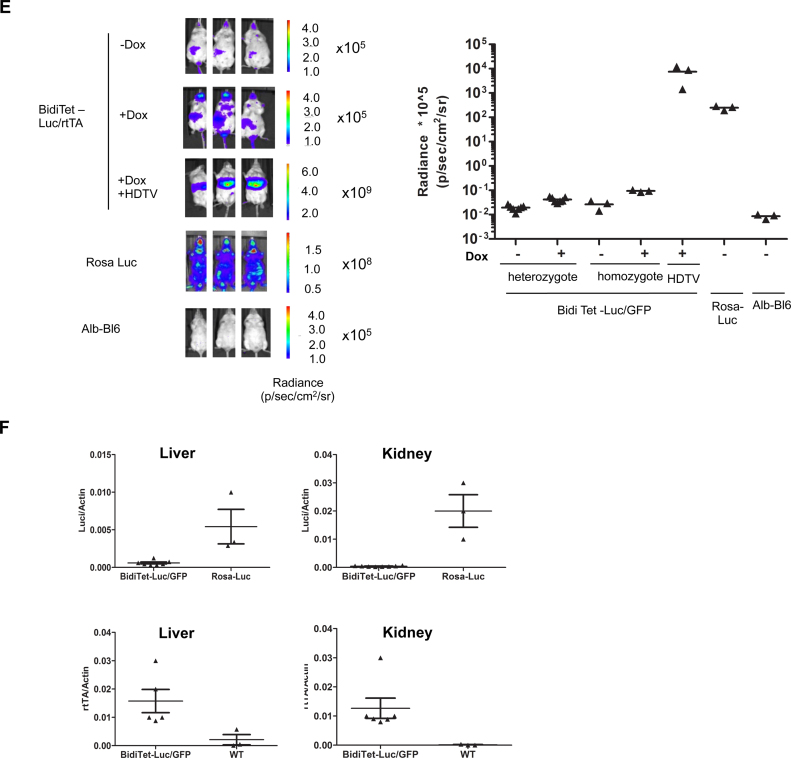
Expression from the bidirectional Tet promoter in the Rosa26 locus is silenced in ES cells, upon differentiation and in transgenic mice. (A) Schematic depiction of the RMCE compatible ROSA26 locus in G4B12 ES cells according to (37) and targeted expression cassettes used in this study. The RMCE strategy relies on Flp mediated targeting of the cassettes which are flanked with heterospecific FRT-WT and FRT-F5 recombination sites (dark and gray ovals). Upon chromosomal integration, an IRES element followed by an ATG from the incoming targeting cassette enables expression of the neomycin phosphotransferase gene, thereby rendering successfully recombined cells resistant against G418. Upon targeting of BidiTet-Luc/GFP, Tet-GFP and BidiTet-Luc/rtTA cassettes, the transactivator (rtTA) is controlled by the endogenous ROSA26 promoter. In BidiTet-Luc/rtTA a second copy of the rtTA is controlled by the cognate promoter, resulting in its positive feedback activation. Tet-GFP is a unidirectional Tet promoter driven GFP cassette. Targeting of a promoter-free GFP cassette resulted in Rosa-GFP cells in which GFP is controlled by the ROSA26 promoter. Rosa-Luc cells were derived from G4B12 ES cells upon stable expression of Cre recombinase. (B) Reporter gene expression in targeted ES cells. ES cells carrying the indicated cassettes were cultivated for 48h in the presence and absence of Doxycycline and analysed for expression of luciferase (as relative light units (rlu) per μg of total protein, left) or GFP (as % GFP expressing cells, right). As a negative control untargeted G4B12 ES cells were included. Cumulative data from two to three independent experiments (each performed in duplicates or triplicates) are represented. Representative flow cytometry plots are indicated for the different cell lines. (C) Stability of expression during culture. The BidiTet-Luc/GFP ES cells were cultured in the presence and absence of Doxycycline. Cells were analysed for luciferase and GFP expression early (passage 2–5) and late (passages 17–20) after targeting. Represented data were derived from two independent experiments in triplicate cell cultures. (D) Stability of reporter expression upon differentiation of BidiTet-Luc/GFP ES cells. To induce differentiation, BidiTet-Luc/GFP ES cells were cultivated on gelatinised plates without LIF in the absence and presence of Doxycycline for 4 and 7 days and subsequently analysed for GFP or luciferase expression. Data were generated from triplicates derived from two (luciferase) or one (GFP) experiments. (E) In vivo imaging of BidiTet-Luc/GFP, Rosa-Luc and wild type albino C57Bl/6 (alb-Bl/6) mice. BidiTet-Luc/GFP mice were analyzed for whole body in vivo bioluminescence upon providing 2 μg/ml Doxycycline in the drinking water for 7 days. BidiTet-Luc/GFP animals that received no Doxycycline were used as controls. BidiTet-Luc/GFP mice are also depicted 2 days after HDTV of the BidiTetLuc/GFP plasmid in the presence of Doxycycline. Note that the scale is different in the groups to accommodate different expression levels. Quantification is depicted on the right. Data from independent mice per group are depicted. (F) To assess the mRNA levels of luciferase and rtTA liver and kidney tissues of Doxycycline induced animals presented in E were harvested. Samples from three to seven animals per group were analysed by qRT-PCR. The levels of luciferase and rtTA transcripts were calculated relative to actin mRNA levels.
AutoTet-Luc/rtTA comprises an autoregulated, bidirectional Tet cassette for controlled expression of luciferase and rtTA in a bicistronic expression unit together with the neomycin resistance gene.
The rtTA expressing vector has been recently described (pCMVRTA2Hyg in (38)). It comprises the CMV promoter driving the rtTA2-S2 transactivator (39) and a hygromycin resistance gene linked by an EMCV-IRES element. TET1c-rtTA was generated from rtTA by including the catalytic domain from the mouse TET1 protein which is fused to the C terminus of the rtTA gene via a XhoI cutting site (CTC/GAG) and a GGA (Glycin) / TCT (Serin) linker. TET1c comprises only the catalytic domain of TET1 lacking any DNA binding domains in the same plasmid background. pCAGGTet1crtTA is a lentiviral vector encoding TET1c-rtTA and dsRed genes in a bicistronic message controlled by the CAGG promoter. Maps and vector sequences are available on request.
Targeted integration by recombinase mediated cassette exchange (RMCE)
For ES cell targeting, 1 × 104 G4B12 cells were seeded in the 6 well plate without feeders. The cells were cotransfected with the FLPe expression vector pFlpe (40) and the respective targeting vectors (BidiTet-Luc/GFP, Tet-GFP, BidiTet-Luc/rtTA or Rosa-GFP) using Lipofectamine 2000 (Life technologies) according to manufacturer's protocol. Media was exchanged 5 h after transfection. 24 h post transfection, the ES cells were transferred to feeder coated 10 cm cell-culture dishes. Selection pressure was added 48 h after transfection (0.4 mg/ml G418). Non-transfected ES cells and ES cells transfected with only the Flp recombinase expression plasmid were routinely included as negative controls. RMCE targeted, G418 resistant cells were then cultured, expanded and analysed for GFP and Luciferase expression.
Gene expression analysis
Total RNA was prepared from ∼1 × 106 cells using the RNeasy Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. The RNA samples were eluted in 30–50 μl RNase-free H2O. Quality and quantity of the isolated RNA samples were assessed using the NanoDrop ND-100 spectrophotometer. Reverse transcription was carried out using the Ready-To-Go First Strand Beads (GE Healthcare) and OligodT primers (Qiagen) according to the manufacturer's instructions. Briefly, a volume corresponding to 5 μg of eluted RNA was denatured at 65°C for 10 min, followed by 5 min incubation on ice and then mixed with OligodT primers and transferred to the Ready-To-Go First Strand Beads for reverse transcription at 37°C for 1 h. 1:10 diluted cDNAs were used as a template for amplification with specific primers for coding sequences of rtTA (rtTA1: 5′-GCATCATACCCACTTCTGCC, rtTA2 5′-GGCCCACGGCGGACAGAGCG) and luciferase (Lucfwd: 5′-GGTGTTGGAGCAAGATGGAT; Lucrev: 5′-TCAAAGAGGCGAACTGTGTG).
For real-time PCR the following components were mixed: 10μl SYBR green (Qiagen) RT-PCR mix, 1 μl forward primer (10pmol), 1μl reverse primer (10pmol), 8 μl cDNA. Real-time PCR was performed on a LightCycler 480 apparatus (Roche). The housekeeping gene Actin (Actinfwd: 5′-TGGAATCCTGTGGCATCCATGAAAC Actinrev: 5′-TAAAACGCAGCTCAGTAACAGTCCG) was used for normalization. Reactions were performed in duplicates or triplicates.
Flow cytometry analysis
For flow cytometry, cells were detached from adherent culture by enzymatic treatment and resuspended in a buffer (2% FBS in PBS). Cells were analyzed using Becton-Dickinson FACScalibur.
In vitro luciferase measurement
The cells were washed in PBS and harvested in 200 μl RLB Buffer (Promega, E397A). 20 μl of this solution was mixed with 100μl detection solution (Beetle Lysis Juice, PJK) and measured using the luminometer Lumat LB 9507 (Berthold). Light units obtained from 10 s were related to the protein content. The protein content was measured with Nanodrop biophotometer (Eppendorff).
Bisulfite Sequencing of the Tet promoter
The high molecular weight DNA was isolated from 1 × 106 cells using the Qiaamp DNA Mini Kit (51304). 300–900 ng of the isolated DNA was used for bisulfite sequencing using the EZ DNA methylation kit (Zymo cat #.D5002 or D5001) according to the manufacturer's instructions. The Tet promoter sequence was amplified from the converted DNA using primers T1 5′-GGTTTTGGTGTTTTTTATGGAGGTTAAAAT-3′ and T2 5′-AACTCTACTTATATAAACCTCCCACC-3′. The reaction was performed on a PCR machine (Biometra). DNA fragments from ES cells and the animals were amplified with: (a) 15 min/95°C, (b) 30 s/95°C, 60 s/55°C, 60 s/72°C (45 cycles), (c) 7 min/72°C, (d) final hold 4°C. Fragments from the immortalized fibroblasts were amplified using: (a) 5 min/95°C, 10 min/72°C, (b) 30 s/95°C, 45 s/56°C, 90 s/72°C (31 cycles), (c) 10 min/72°C, (d) final hold 16°C.
Amplified PCR products were integrated into the PCR blunt cloning vector (Invitrogen) using the protocol according to the manufacturer's instructions. Upon transformation, independent clones were picked and expanded. Plasmid DNA was isolated, purified and sequenced (HZI sequencing facility). Sequences were analysed for the presence of C to T conversions indicating unmethylated CpGs.
Epityper analysis
For EpiTYPER analysis DNA was converted using the EZ DNA methylation kit (Zymo) according to the manufacturer's protocol. Briefly, 1 μg of genomic DNA in 50 μl 1× M-Dilution Buffer was mixed with 100 μl of CT Conversion Reagent and incubated in a PCR machine 30 s/95°C and 15 min/50°C (20 cycles) and then cooled down. After adding 400 μl of M-Binding Buffer the sample was loaded on a Zymo-Spin I Column, washed, desulfonated, washed and eluted in 100 μl H2O. PCR reactions were performed using 0.5 μl 10× HotStarBuffer, 200 μM dNTPs, 0.2 U Hot Star Taq polymerase and 5–10 ng template DNA, 500 pM primers (ROSA26: P1 5′-TTTGGGAGAGTAAGTGTTTTTTGTTTTTAA-3′ and P2 5′-ACCCTCCCCTTCCTCTAAAAAAATC-3′ and (Thumpd3: P1 5′-GGGGGAGAAGAATTTTTTTAGTTT-3′ and P2 5′-ATAACTCTCTCACTCCCACCCTCTAC-3′) in a final reaction volume of 5 μl. PCR was performed in a Veriti 384 well thermal cycler (Applied Biosystems) with the following steps: 94°C/4 min; 45× [94°C/20 s; 30 s/59°C; 60 s/72°C]; 3 min/72°C.
Unincorporated nucleotides were removed by shrimp alkaline phosphatase prior to in vitro transcription with T7 RNA polymerase, which is guided to the amplified PCR-products by the introduced T7 promoter tag in the reverse primer. The transcribed RNA was enzymatically cleaved at T using RNaseA according to the manufacturer's instruction and desalted. The DNA samples are transferred on a SpectroCHIP with the Phusio Chip Module and analysed with the MassARRAY Compact System MALDI-TOF MS (all Sequenom). Acquired data was processed with the EpiTyper Analyzer software (version 1.0.5, Sequenom).
Mice
BidiTet-Luc/GFP targeted murine embryonic stems cells were used to generate transgenic mice by blastocyst injection. Rosa-Luc mice were previously described as ROSAConL mice (37). AutoTet-Luc/rtTA mice were obtained from 129 ES cells upon random integration of AutoTet-Luc/rtTA. All mouse lines were used upon backcrossing to albino C57Bl/6 to achieve white coat colour for increased sensitivity of bioluminescence measurements. Mice were maintained in the breeding facilities of Helmholtz Centre for Infection Research (HZI, Braunschweig, Germany) using individually ventilated cages. All animal experiments were carried out in accordance with national and local regulations.
Doxycycline was administered to mice via supplementing the drinking water at a final concentration of 2mg/ml. 2.5 mg/kg of 5-Azacytidine (Sigma, dissolved in PBS and stored at –20°C) administered i.p. on two consecutive days (d0 and d1).
For hydrodynamic tail vein injection (HDTV) 25 μg of the respective plasmids were dissolved in PBS (10% of body weight) and injected into the tail vein of the animals within 5–10 s.
For bioluminescence Xenogen IVIS 200 was used. Animals were anaesthetized in the induction chamber by 2–2.5% isoflurane (Albrecht). Mice were then injected intraperitoneally with 100 μl of luciferin (30 mg/ml in PBS, Synchem OHG) and placed on the heated (37°C) platform in the acquisition chamber. Anesthesia was maintained by constant administration of isoflurane via nose cones while images were taken. Initially, a grey-scale image was taken in the light tight chamber. Photons were collected by a sensitive CCD camera and the signals were overlaid to the grey-scale image. Analyses of images was done with the Living image 2.60.1 (Igor Pro 4.09A) computer program.
For histological analysis of liver, mice were sacrificed and livers were fixed for 48h in formalin and then dehydrated in 70% ethanol before processing for Hematoxylin & Eosin (H&E) and GFP staining by the histopathology facility of the HZI.
Statistical analysis
The data are displayed as mean and standard deviation. The significance was calculated by Mann-Whitney U test for all comparisons (GraphPad Prism 5.04, GraphPad Software, La Jolla, CA, USA). Differences between the groups were considered as follows: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001
RESULTS
The Tet promoter is silenced upon integration into the ROSA26 locus
We established murine ES cells with different Tet-promoter dependent reporter gene expression cassettes (Figure 1A) by recombinase mediated cassette exchange (RMCE). The cassettes were integrated downstream of the ROSA26 promoter (37) in RMCE compatible G4B12 cells (33). Bidirectional (BidiTet-Luc/GFP) and unidirectional (Tet-GFP) Tet promoter cassettes encoding GFP and/or luciferase were constructed. In these cassettes, the rtTA (Tet-on) transactivator gene is controlled by the host's ROSA26 promoter. As controls, we used targeted cells in which the coding sequences of luciferase and GFP are driven by the ROSA26 promoter resulting in constitutive expression (Rosa-Luc, Rosa-GFP).
Populations of targeted cells were induced by Doxycycline for two days and analysed for luciferase expression (Figure 1B). In BidiTet-Luc/GFP cells, luciferase expression was induced by Doxycycline resulting in an average expression of 2.6 × 103 RLU/μg of total protein. In Rosa-Luc cells, in which luciferase is controlled by the endogenous ROSA26 promoter, a 15-fold higher expression could be observed. We evaluated if Doxycycline induced expression could be enhanced by installing a positive feedback loop. Unexpectedly, BidiTet-Luc/rtTA cells, in which a second rtTA coding sequence is controlled by the Tet promoter, showed a similar luciferase expression pattern when compared to BidiTetLuc/GFP cells, indicating that levels of rtTA molecules are not the limiting factor for expression (Figure 1B).
We determined the frequency of GFP expressing BidiTet-Luc/GFP cells as well as Tet-GFP cells. If compared to luciferase, detection of GFP requires a higher threshold expression level, however, it provides the option to monitor expression of individual cells by flow cytometry. Interestingly, in the presence of Doxycycline, GFP expression was induced in 85% of the Tet-GFP cells while ∼20% of the BidiTet-Luc/GFP cells showed GFP expression, albeit at a lower level. In contrast, we observed homogeneous GFP expression in virtually all cells when GFP is transcribed from the endogenous ROSA26 promoter as in ROSA-GFP cells (Figure 1B). The heterogeneous, mosaic type of expression in individual BidiTet-Luc/GFP and Tet-GFP cells suggests that the Tet promoter is susceptible to epigenetic modulations in the ROSA26 locus, leading to a decrease in expression strength down to total inactivity in a large portion of the population.
To investigate stability of expression over time, we cultured BidiTet-Luc/GFP cells for 20 passages in the presence or absence of Doxycycline and subsequently measured expression of luciferase and GFP. When compared to cells in early passages, luciferase expression in the presence of Doxycycline decreased ∼14-fold and GFP expression was found in <2% of cells (Figure 1C). To investigate if expression is altered upon differentiation the cells were cultured in absence of LIF and reporter gene expression was assessed over time. Intriguingly, on day 7, when cells had lost stem cell phenotype as indicated by absence of alkaline phosphatase (Supplementary Figure S2) GFP expression was not detectable and luciferase expression was decreased (Figure 1D) indicating extensive silencing.
To evaluate Doxycycline dependent gene expression in vivo we established a transgenic mouse line from BidiTet-Luc/GFP ES cells. Expression in individual animals was assessed by whole body bioluminescence imaging. As controls, Rosa-Luc mice were used in which luciferase is controlled by the ROSA26 promoter (Figure 1A) (37) as well as wild type albino C57Bl/6 mice. Untreated BidiTet-Luc/GFP mice showed low basal expression of luciferase which was only increased ∼2-fold when mice were induced with Doxycycline for 7–11 days (Supplementary Figure S3A). The signal was approximately four orders of magnitude lower than in ROSA-Luc mice (Figure 1E). Breeding animals in the presence of Doxycycline had no significant effect on the expression level (Supplementary Figure S3B). Even in homozygous BidiTet-Luc/GFP animals with two copies of the inducible expression cassette Doxycycline dependent expression was low (Figure 1E). We tested if expression can be achieved by de novo introduction of the BidiTet-Luc/GFP cassette in liver tissue of mice. To this end, we performed hydrodynamic tail vein (HDTV) injection of the BidiTet-Luc/GFP plasmid into BidiTet-Luc/GFP mice which should mainly create an episomal status of the cassette in liver cells (41). On day 2 after HDTV injection, high luciferase expression was observed confirming the functionality of the Tet cassette (Figure 1E). This suggests that in the BidiTet-Luc/GFP animals the integrated cassette is strongly silenced.
To determine transgene expression in individual organs, we sacrificed Doxycycline induced and non-induced BidiTet-Luc/GFP animals and measured RNA levels of luciferase and rtTA in liver and kidney tissue. In both organs, the RNA levels of luciferase in Doxycycline induced BidiTet-Luc/GFP animals were lower compared to those in Rosa-Luc animals (Figure 1F). We found significant rtTA expression in these organs, indicating that the endogenous ROSA26 promoter is active. Together, these observations show that the Tet promoter driven expression cassettes are susceptible to silencing in vitro as well as in transgenic animals.
The Tet promoter but not the neighbouring host cell promoters becomes methylated
To investigate if DNA methylation could contribute to the silencing of Tet cassettes in the ROSA26 locus we treated Doxycycline-induced and mock-induced BidiTet-Luc/GFP ES cells with inhibitors of DNA methylation (DNMTi). Upon cultivation of cells in the presence of Decitabine, the number of GFP expressing ES cells could be increased from 2% in non-treated cells to about 6% and 8% in treated cells, respectively (Figure 2A). Also, luciferase expression was increased in a dose dependent manner (Supplementary Figure S1C). This suggests that DNMTi can restore expression of the Tet cassette in a fraction of ES cells.
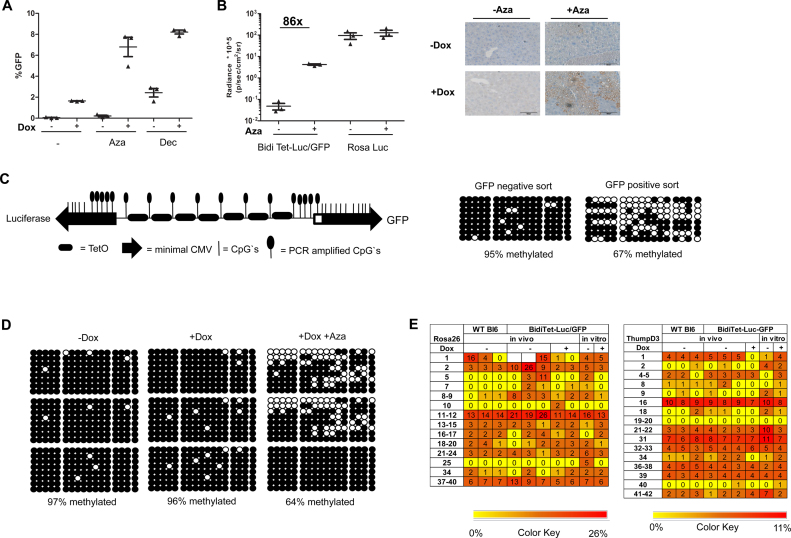
Figure 2.
The bidirectional Tet promoter is methylated in the ROSA26 locus and can be reactivated by DNMTi (A) GFP expression in BidiTet-Luc/GFP ES cells upon cultivation in presence of the DNMTis Decitabine and Azacytidine. BidiTet-Luc/GFP ES cells were cultivated in presence or absence of Doxycycline and Decitabine or Azacytidine for three days and subsequently subjected to flow cytometry. The percentage of GFP expressing cells is depicted. (B) In vivo imaging of induced BidiTet-Luc/GFP mice upon treatment with Azacytidine. Whole body bioluminescence was determined in three independent animals before and three days after the first treatment with Azacytidine. As control, Rosa-Luc animals were used. The bioluminescence of three independent mice per group is depicted on the left. On day 3, the liver was isolated and slices were stained for GFP expression. Depicted are representative sections from mice of the indicated groups. (C) Schematic depiction of the bidirectional Tet promoter in BidiTet-Luc/GFP cells and the position of the CpG's. The bidirectional Tet promoter comprises 30 CpGs, 8 CpG's flank the tetO sequences, 3 CpGs are located in a spacer element and the remaining 19 CpG are in the two opposing CMV promoter sequences. The location of the CpGs within the promoter (lines) and within the amplified PCR fragment (lollipops) are indicated. Note that the 5′ minimal CMV promoter is slightly shorter than the 3′ CMV promoter (72). (D) Methylation status of the Tet promoter in BidiTet-Luc/GFP ES cells. Cells were treated for 3 days with Decitabine. GFP positive and negative cells were sorted, DNA was extracted and subjected to bisulfite analysis. Subsequently, the Tet promoter DNA was PCR amplified and cloned in E. coli. Sequences of eight randomly picked clones representing eight independent cells are depicted. In each line, the circles depict the 17 CpGs of an individual clone/cell. Black circles indicate methylated cytosines and non-filled circles non-methylated cytosines. (E) Methylation status of Tet promoter in BidiTet-Luc/GFP mice. Bisulfite analysis of the Tet promoter based on DNA extracted from mouse livers. The methylation status of the Tet promoter in the non-induced (–Dox), induced (+Dox) and Azacytidine/Dox induced animals (+Dox +Aza) is depicted. Three independent mice per group were analysed. For each mouse, eight independent cells were sequenced and analysed. Black circles indicate methylated cytosines and non-filled circles non methylated cytosines. (F) Epityper based DNA methylation analysis of the ROSA26 and ThumpD3 promoter in cells and mice. The heat maps represent the results of the methylation analysis done on Epityper. The various CpG motifs of the two promoters are indicated on the x-axis of each map and the samples used are indicated on the y-axis. The reference scale is indicated (0.01 = 1% methylation and 0.26 = 26% methylation). Each vertical column represents the same CpG motif analyzed from different gDNA samples while each row represents the different CpGs within the same sample.
We also examined the role of DNA methylation for silencing Tet driven transgene expression in vivo. BidiTet-Luc/GFP mice were treated with Azacytidine for two consecutive days and subsequently evaluated by in vivo luminescence. Administration of Azacytidine to induced BidiTet-Luc/GFP animals increased expression of luciferase 86-fold. Luciferase activity in Rosa-Luc mice did not change by this treatment (Figure 2B). We also evaluated transgene expression in liver sections of Azacytidine treated and non-treated BidiTet-Luc/GFP mice by staining with a GFP-antibody. Positive staining confirmed activation of GFP expression in individual hepatocytes of DNMTi treated animals (Figure 2B).
To analyse the DNA methylation status of the Tet promoter, GFP positive and negative fractions from DNMTi and Doxycycline treated BidiTet-Luc/GFP mES cells were separated by FACS, genomic DNA was isolated and converted with bisulfite. The bidirectional Tet promoter comprises 30 CpGs (Figure 2C). PCR amplification of the entire promoter region is challenging, presumably due to the high degree of sequence repeats. Thus, we restricted our analysis to a PCR fragment comprising 17 CpGs, in particular the 8 CpGs located around the tetO sequences, 5 CpGs in the luciferase driving minimal CMV promoter and 4 CpGs in the 5′ region of the GFP driving CMV promoter (Figure 2C). The fragments were cloned in Escherichia coli. Eight bacterial clones reflecting the promoter sequence of independent ES cells were randomly picked and sequenced.
Interestingly, the methylation analysis revealed drastic differences in the overall methylation level of expressing vs. non-expressing cells. In GFP negative ES cells, nearly all CpGs in the amplified promoter region were methylated (Figure 2C). In contrast, in the GFP expressing cells, the level of methylation was reduced to 67%. Together, this confirms an inverse correlation between the transgene expression level and the degree of bidirectional Tet promoter methylation. We also investigated the methylation status of the Tet promoter in the liver from BidiTet-Luc/GFP mice in the presence and absence of Doxycycline. >95% of CpGs within the Tet promoter were methylated irrespective of Doxycycline treatment. Importantly, if the mice were subjected to DNMTi treatment the overall methylation level of the Tet promoter was decreased to about 64% (Figure 2D).
We wondered about the degree of methylation of the endogenous ROSA26 locus and if this would be altered by integration of the Tet-transgene cassettes. To elucidate this, we analysed the DNA methylation status of the cellular ROSA26 and ThumpD3 promoters adjacent to the transgenic Tet cassette in BidiTet-Luc/GFP mice (Figure 1A). Since both host promoter sequences are devoid of repetitive elements they were subjected to Epityper analysis, which provides quantitative data relying on a large number of reads/data points (42,43). In vitro methylated plasmid DNA served as a control. The DNA methylation analysis revealed very low methylation levels for both, ThumpD3 and ROSA26, promoters and no significant alteration by transgene integration and subsequent treatments and cultivation (Figure 2E and Supplementary Figure S4). This indicates that the high methylation level of the synthetic Tet promoter does not spread to the neighboring host promoters. These results rule out that the methylated status of the synthetic Tet promoter modulates expression of the ROSA26 promoter.
Re-activation of the silenced Tet promoter by targeted demethylation
We asked if methylation induced silencing could be reverted by targeting the demethylation domain of ten eleven translocation protein 1 (TET1) to the Tet promoter. To this end, we generated a gene that encodes a fusion protein in which the catalytic domain of TET1 is linked to the carboxy terminus of the rtTA transactivator. This fusion protein is designated TET1c-rtTA in the following.
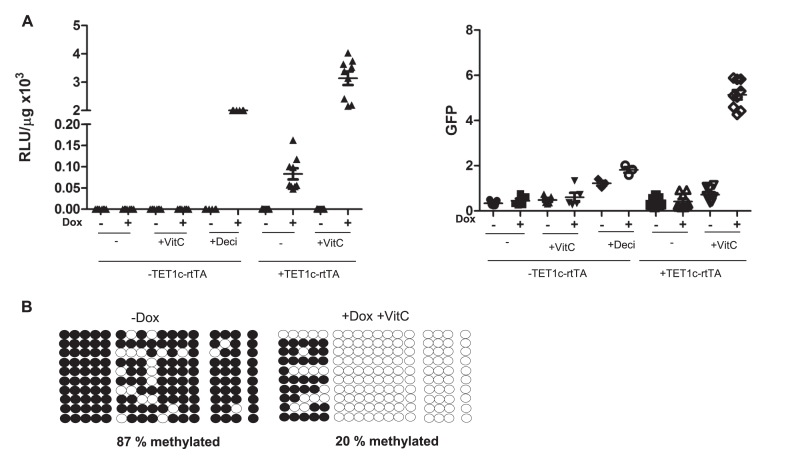
To investigate the potential of TET1c-rtTA to overcome the methylation of the Tet promoter we stably integrated a CAGGS promoter driven TET1c-rtTA cassette into immortalized BidiTet-Luc/GFP fibroblast cells. We analysed expression in the presence of Vitamin C, which is known as a co-factor of TET proteins (44). In agreement with previous results, in absence of TET1c-rtTA BidiTet-Luc/GFP fibroblasts did not show Doxycycline dependent luciferase expression. In BidiTet-Luc/GFP fibroblasts that additionally expressed TET1c-rtTA, luciferase and GFP expression were found to be increased (Figure 3A and B and Supplementary Figure S5). Of note, the enhancing effect of TET1c-rtTA was strictly dependent on the presence of Doxycycline and the cofactor Vitamin C (Figure 3B). Absence of either Doxycycline or Vitamin C led to loss of expression, indicating that both factors are critically needed. We can exclude that endogenous TET1 proteins activate expression upon binding to the methylated Tet promoter since treatment of BidiTet-Luc/GFP fibroblasts with Vitamin C alone did not induce luciferase expression (Figure 3A).
Figure 3.
The silenced Tet promoter can be reactivated by site specific targeted demethylation in fibroblasts. (A) Reactivation of silenced reporter expression in fibroblasts. Immortalized fibroblasts from BidiTet-Luc/GFP mice were transduced with Tet1c-rtTA by lentiviral gene transfer. Infected (+TET1c-rtTA) and non-infected (–TET1c-rtTA) cells were cultured with or without Doxycycline, Vitamin C (+VitC) for three days as indicated and subsequently analysed for luciferase and GFP expression. As control, the non-transduced BidiTet-Luc/GFP fibroblasts were cultured with Decitabine. The data from two or three independent experiments with duplicates or triplicates are depicted. (B) Methylation status of the Tet promoter in Tet1c-rtTA transduced fibroblasts. The genomic DNA was isolated from TET1c-rtTA transduced BidiTet-Luc/GFP fibroblasts previously cultured in absence or presence of Doxycycline and Vitamin C. The DNA was converted with bisulfite. The Tet promoter DNA was PCR amplified, cloned and sequenced. The methylation status of 10 independent clones is depicted. Black circles indicate methylated cytosines and non-filled circles non-methylated cytosines.
To confirm that expression of the TET1c-rtTA facilitates increased expression by reverting the methylation status of the Tet promoter, the DNA of TET1c-rtTA transfected BidiTet-Luc/GFP cells was subjected to methylation analysis. Only 20% of the Tet promoter CpGs were found to be methylated in the presence of Vitamin C and the fusion protein while nearly 90% of CpGs were methylated in the absence of the fusion protein irrespective of Vitamin C (Figure 3B and Supplementary Figure S6). This shows that TET1c-rtTA, but not endogenous TET proteins rescue silenced expression from the BidiTet promoter by supporting its demethylation.
Targeted demethylation reactivates silenced Tet cassettes in mice
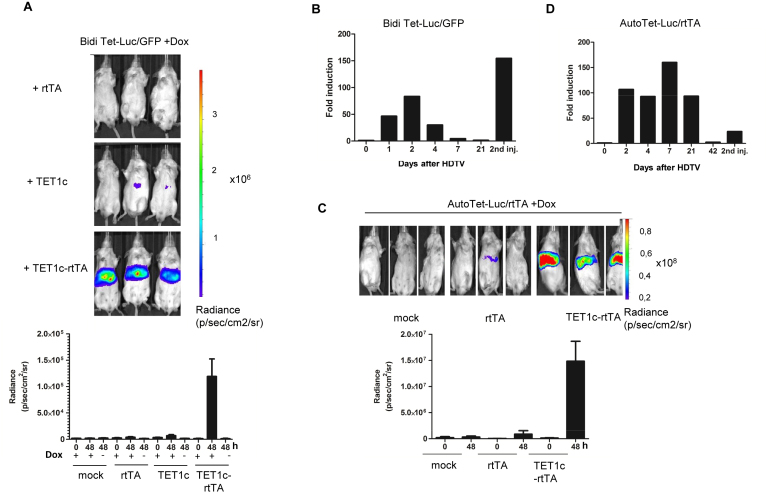
Finally, we investigated if expression can be reactivated in vivo. Since Vitamin C (ascorbate) is synthesized from glucose in mouse liver (reviewed in (45)) no additional supplementation was required. We injected the TET1c-rtTA expression plasmid into Doxycycline treated BidiTet-Luc/GFP animals by HDTV injection. As controls, we employed plasmids encoding for rtTA or the catalytic domain of TET1 (TET1c) lacking the DNA binding domain. The animals were analysed for bioluminescence 48h after injection. None of the control groups showed a significant increase in luciferase expression suggesting that neither the injection itself nor the elevated expression of rtTA or the non-binding catalytic domain could significantly increase luciferase expression (Figure 4A). However, upon administration of TET1c-rtTA the animals showed a pronounced upregulation of luciferase (about 80fold). Of note, upregulation was dependent on TET1c-rtTA binding to the Tet promoter since in absence of Doxycycline no increase in expression was observed (Figure 4A). In line with this, when we injected TET1c-rtTA in absence of Doxycycline no significant increase in expression could be observed while subsequent administration of Doxycycline rescued expression (Supplementary Figure S7).
Figure 4.
Reversion of Tet-promoter silencing in vivo. (A) Reactivation of silenced gene expression in vivo. BidiTet-Luc/GFP mice were subjected to HDTV using the indicated plasmid DNA. As negative control the animals were hydrodynamically injected with PBS (mock). After 48 h, bioluminescence was measured by in vivo imaging. Doxycycline feeding was started 7 days before the experiment and was maintained throughout the experiment (upper panel). Control groups were measured in absence of Doxycycline as indicated. Representative in vivo images of mice as well as cumulative data from 5 independent animals per group are shown (lower panel). (B) Kinetics of reactivation. Bioluminescence of BidiTet-Luc/GFP animals was monitored on the indicated days after HDTV injection as described in (A). On day 21, HDTV injection was repeated using the same plasmid DNA and animals were imaged one day later. Bars represent measurement of four animals. The fold induction is related the bioluminescence signal before HDTV injection. Day 2 luciferase measurements were derived from the experiment represented in (A). (C) Reactivation of silenced expression in AutoTet-Luc/rtTA mice. Doxycycline induced AutoTet-Luc/rtTA mice received TET1c-rtTA, rtTA or PBS (mock) via HDTV injection. Bioluminescence of mice 48 h after HDTV injection is depicted on the left. The quantification of bioluminescence before (0) and 48 h after (48) based on three independent animals is depicted in the lower panel. As negative control (mock) AutoTet-Luc/rtTA animals were injected with PBS. (D) Kinetics of reactivation in AutoTet-Luc/rtTA mice. Bioluminescence of AutoTet-Luc/rtTA mice of (C) was monitored on the indicated days after HDTV injection. AutoTet-Luc/rtTA animals received drinking water supplemented with Doxycycline throughout the experiment. The animals were measured on the indicated days before and after HDTV injection. On day 42 HDTV injection was repeated using the same plasmid DNA and animals were analysed one day later. Bars represent measurement of three animals. Day 2 luciferase measurements are derived from the experiment represented in (C). The fold induction is related to the bioluminescence before HDTV injection.
We asked if the silencing activity of ROSA26 chromosomal environment on Tet cassettes is restricted to the course of embryonic development or if it would also dominate expression of Tet cassettes after differentiation, i.e. in adult animals. Thus, we investigated if expression of Tet promoter, once actively demethylated by expression of TET1c-rtTA, would be maintained in absence of TET1c-rtTA. We exploited the transient expression of HDTV transferred plasmid DNA in liver (46) and followed expression of these treated animals over three weeks. Doxycycline was continuously provided during the course of the experiment. The analysis showed a peak of luciferase expression around day 2 after HDTV. Then, expression continuously decreased and was comparable to silenced expression levels after 3 weeks. When we repeatedly injected TET1c-rtTA plasmid, luciferase expression increased to levels comparable to the first injection (Figure 4B) while injection of control plasmids did not result in significant changes (Supplementary Figure S8A and B). Together, this indicates that long-term activation of the silenced Tet promoter in the ROSA26 locus requires permanent demethylation by TET1c-rtTA.
We investigated if reactivation of silenced expression could be also achieved for another chromosomal integration site. To this end, we generated AutoTet-Luc/rtTA ES cells by random integration of a single copy of an autoregulated bidirectional Tet cassette and also established transgenic mice thereof. While expression of AutoTet-Luc/rtTA ES cells in the presence of Doxycycline was comparable to ROSA-Luc ES cells (Supplementary Figure S9A) it was found to be 20-fold decreased on the level of mice suggesting silencing of expression during development (Supplementary Figure S9B). When we injected TET1c-rtTA into these animals, we observed a drastic increase in expression while injection of the control rtTA plasmid did not result in significant changes (Figure 4C; Supplementary Figure S10). Interestingly, also in these mice the effect of TET1c-rtTA was transient but declined less rapidly. Similar to the results with the BidiTet-Luc/GFP mice, a second injection with TET1c-rtTA could rescue luciferase expression (Figure 4D).
Together, the results show that Tet promoters are prone to epigenetic silencing and that targeted demethylation of the Tet promoter is a tool to selectively reactivate silenced expression in a transient manner in vitro and in vivo, independent of the integration site.
DISCUSSION
The intensity of transgene expression is considered to be affected by both, the composition of the regulatory promoter/enhancer elements driving the gene of interest and the chromosomal site of integration (47). These factors not only determine the initial transgene expression level, they also affect the stability of transgene expression. Indeed, while the expression level in certain integration sites is found to be stable over extended cultivation periods even when perturbed by external alterations of culture conditions, in many integration sites, transgene expression underlies changes, mostly inactivation. While screening methods today allow measurement of the level of initial transgene expression and thus the isolation of relevant cell clones in high throughput, the extent and stability of transgene expression needs to be defined for each cell clone and cultivation/environment condition.
Targeted integration into defined chromosomal sites was suggested to overcome the variability associated with random integration of expression cassettes. Accordingly, specific targeting of transgenes into predefined and tagged sites with the help of site specific recombinases such as Flp or Cre (reviewed in (48)) was introduced. Upon targeting a given cassette carrying the same regulatory elements into a defined locus, isogenic subclones display homogenous expression levels indicating that the influence from a previously characterized integration site is maintained constant. Accordingly, the impact on defined expression cassettes can be predicted (36,49,50). This approach has been boosted by methods to enhance the efficiency of homologous recombination. Together with increasing information of sequence and epigenetic status for whole genomes the employment of site specific double stranded DNA breaks by Zn finger nucleases, TALENs, or CRISPR/Cas9 (51) facilitates targeted integration of expression cassettes into virtually any site of the genome. Irrespective of the method used for targeted integration, however, the interaction of various incoming cassettes (with different regulatory elements) with the chromosomal environment is still not predictable and remains to be evaluated empirically. Thus, methods that overcome silencing of transgene cassette are of interest.
We investigated the expression and crosstalk of reporter gene expression cassettes upon targeted integration into the ROSA26 locus. Although this locus has been described to be ubiquitously active (32), expression of transgene cassettes with different transcription control elements underlies non-predictable expression characteristics (52,53). While Tet promoter driven transgene expression could be achieved to a certain degree, mosaic and also loss of expression have been reported (34,54–56). This suggests that the Tet promoter is affected by epigenetic silencing, although the underlying mechanism remained elusive. In the current study, we focused on Doxycycline regulatable cassettes integrated into the ROSA26 site to follow expression levels under various conditions. Targeting of these cassettes into mES cells allowed us to monitor expression during and after differentiation, conditions that are well-known for concerted changes in gene expression patterns. We observed a strong decline of transgene expression i) upon mid-term cultivation of ES cells, ii) during differentiation and iii) after creation of transgenic animals, indicating that the transgene cassettes were epigenetically silenced.
The mechanisms that lead to the induction of silencing are not well understood (47,57). DNA methylation is one of the important mechanisms associated with silencing. An early study suggested that DNA methylation may spread from highly methylated foci to the nearby regions (58). In particular retroviral elements have the capacity to induce de novo methylation of surrounding chromosomal sites (58,59). The spreading of DNA methylation to distal sites was shown to be driven by recruitment of repression complexes to the methylated sites, which in turn can render the chromatin more susceptible to the DNA methylation machinery (60). In our study, we observed that the highly methylated Tet promoter does not induce methylation of the flanking endogenous ROSA26 and ThumpD3 promoter. Thus, both host promoters are not affected by strong methylation induced by the incoming constructs, suggesting that these host promoters are protected from this silencing mechanism. Recently, insulator sequences were utilized to stabilize expression of Tet cassettes (14,15,61). While these elements could increase overall expression, they did not overcome the mosaic type of expression upon differentiation (34).
To overcome silencing we developed a new tool to specifically stabilize transgene expression. The TET1c-rtTA fusion protein demethylates DNA after Doxycycline dependent binding to the Tet operator driving the transgene. We show that expression of the TET1c-rtTA fusion protein can rescue silenced Tet cassettes both in cells as well as in mice (Figures 3A and 4). Of note, due to the Doxycycline dependent demethylation, the induced but not the basal expression of the system is significantly enhanced, thereby increasing the overall induction rate (Figure 4 and Supplementary Figure S9). Targeted demethylation of specific genomic sites was recently approached by site specific recruitment of the enzymatic activity of TET proteins upon fusion with TALE or Cas9 proteins (62–64). When targeting a single site the extent of demethylation as such is limited to a few CpGs (62). Multiplexing is usually required to achieve robust demethylation (65). In the system used here, seven DNA motifs are provided which serve as binding sites for the TET1c-rtTA fusion protein, thereby allowing the recruitment of several molecules. This resulted in efficient demethylation of an area of at least 390 bp which was sufficient to restore expression (Figure 3B). In contrast to the constitutive demethylation strategies via TALEN and CRISPR/Cas9, our strategy is characterized by Doxycycline controlled binding of the fusion protein. Thereby, the transcriptional status can be reversibly modulated. Together with the recently emerged Doxycycline inducible recruitment of gene repressors (66,67), this fusion protein complements the toolbox for kinetic control of epigenetic states. However, given the stochastic nature of Doxycycline induced transcription appropriate analytic tools will be required (38,68,69).
We observed that TET1c-rtTA cannot overcome the mosaic type of reporter expression in individual cells. Analysis of the methylation status revealed that in a certain portion of cells demethylation was achieved. Of note, even global impairment of methylation by Decitabine led to a similar phenotype suggesting stochasticity of the demethylation process or the existence of other mechanisms than methylation stabilizing the silencing of the Tet cassette. It will be interesting to follow the fate of cells that failed to become reactivated.
Recently, Wan et al. (56) reported that expression from Tet promoter in ES cells is sensitive to epigenetic silencing after targeted integration into the Col1A1 locus, a site that was previously shown to support Tet induced expression (70). The authors observed that silencing of the Tet cassette in this locus was dependent on the exposure to Doxycycline during the early developmental stages. The conclusion from this study was that in this locus silencing of the Tet promoter is crucially dependent on its transcription in early developmental stages. In contrast, Wiznerowicz et al. reported about irreversible silencing of randomly integrated Tet cassettes by KRAB mediated repression when transcription of the Tet promoter was impaired during the first days of mouse development (71). For the ROSA26 locus, we did not see significant differences when we targeted the construct in the presence or absence of Doxycycline (data not shown). Further, breeding of BidiTet-Luc/GFP animals in the presence of Doxycycline did not reduce the silencing level of the Tet promoter in the ROSA26 locus (Supplementary Figure S2B). Thus, we conclude that for the ROSA26 locus, transcription from the Tet promoter does not influence the epigenetic status of bidirectional Tet cassettes upon differentiation.
On the whole, this study shows that epigenetic silencing of methylation prone promoter sequences occurs even in transcriptionally active chromosomal sites, but without affecting the neighbouring genes. By specific recruitment of epigenetic modifiers silencing of the synthetic cassettes can be overcome. This opens new perspectives for transgenesis as well as epigenetic control of transcription.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Maria Höxter for cell sorting, the central animal facility at HZI for mouse breeding and the transgenic service unit (TGSM) for generating transgenic mice. L.Z. and S.S. wish to thank the HZI International Graduate School for Infection Research for continuous support. Finally, we are grateful to Zhang Yi, Boston Children Hospital, for providing us the plasmid encoding the TET1 catalytic domain.
Footnotes
Present addresses:
Lisha Zha, University Clinic RIA, Department Immunology, Bern, Switzerland.
Pamela Riemer, Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin, Humboldt-Universität zu Berlin, and Berlin Institute of Health, Institute of Pathology, Laboratory of Molecular Tumor Pathology, Berlin, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG, German Research Foundation): Cluster of Excellence REBIRTH (from Regenerative Biology to Reconstructive Therapy) and SFB900 (Chronic Infection); Helmholtz Association of German Research Centres: cross-program topic ‘Metabolic Dysfunction’; Funding for open access charge: Helmholtz Centre for Infection Research (HZI).
Conflict of interest statement. None declared.
REFERENCES
- 1. May T., Butueva M., Bantner S., Markusic D., Seppen J., MacLeod R.A., Weich H., Hauser H., Wirth D.. Synthetic gene regulation circuits for control of cell expansion. Tissue Eng. Part A. 2010; 16:441–452. [DOI] [PubMed] [Google Scholar]
- 2. Saxena P., Heng B.C., Bai P., Folcher M., Zulewski H., Fussenegger M.. A programmable synthetic lineage-control network that differentiates human IPSCs into glucose-sensitive insulin-secreting beta-like cells. Nat. Commun. 2016; 7:11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hockemeyer D., Soldner F., Cook E.G., Gao Q., Mitalipova M., Jaenisch R.. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008; 3:346–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Sudhof T.C., Wernig M.. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010; 463:1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pfisterer U., Kirkeby A., Torper O., Wood J., Nelander J., Dufour A., Bjorklund A., Lindvall O., Jakobsson J., Parmar M.. Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:10343–10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Karlsson M., Weber W., Fussenegger M.. Design and construction of synthetic gene networks in mammalian cells. Methods Mol. Biol. (Clifton, N.J.). 2012; 813:359–376. [DOI] [PubMed] [Google Scholar]
- 7. Xie M., Fussenegger M.. Mammalian designer cells: engineering principles and biomedical applications. Biotechnol. J. 2015; 10:1005–1018. [DOI] [PubMed] [Google Scholar]
- 8. Auslander S., Fussenegger M.. Engineering gene circuits for mammalian cell-based applications. Cold Spring Harb. Perspect. Biol. 2016; 8:a023895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gossen M., Bujard H.. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gossen M., Freundlieb S., Bender G., Muller G., Hillen W., Bujard H.. Transcriptional activation by tetracyclines in mammalian cells. Science (New York, N.Y.). 1995; 268:1766. [DOI] [PubMed] [Google Scholar]
- 11. Zhu Z., Zheng T., Lee C.G., Homer R.J., Elias J.A.. Tetracycline-controlled transcriptional regulation systems: advances and application in transgenic animal modeling. Semin. Cell Dev. Biol. 2002; 13:121–128. [DOI] [PubMed] [Google Scholar]
- 12. Carey B.W., Markoulaki S., Beard C., Hanna J., Jaenisch R.. Single-gene transgenic mouse strains for reprogramming adult somatic cells. Nat. Methods. 2010; 7:56–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matsunaga T., Yamashita J.K.. Single-step generation of gene knockout-rescue system in pluripotent stem cells by promoter insertion with CRISPR/Cas9. Biochem. Biophys. Res. Commun. 2014; 444:158–163. [DOI] [PubMed] [Google Scholar]
- 14. Haenebalcke L., Goossens S., Dierickx P., Bartunkova S., D’Hont J., Haigh K., Hochepied T., Wirth D., Nagy A., Haigh J.J.. The ROSA26-iPSC mouse: a conditional, inducible, and exchangeable resource for studying cellular (De)differentiation. Cell Rep. 2013; 3:335–341. [DOI] [PubMed] [Google Scholar]
- 15. Hsiao E.C., Nguyen T.D., Ng J.K., Scott M.J., Chang W.C., Zahed H., Conklin B.R.. Constitutive Gs activation using a single-construct tetracycline-inducible expression system in embryonic stem cells and mice. Stem Cell Res. Ther. 2011; 2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dow L.E., Fisher J., O’Rourke K.P., Muley A., Kastenhuber E.R., Livshits G., Tschaharganeh D.F., Socci N.D., Lowe S.W.. Inducible in vivo genome editing with CRISPR-Cas9. Nat. Biotechnol. 2015; 33:390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Premsrirut P.K., Dow L.E., Park Y., Hannon G.J., Lowe S.W.. Creating transgenic shRNA mice by recombinase-mediated cassette exchange. Cold Spring Harbor Protoc. 2013; 2013:835–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dow L.E., Premsrirut P.K., Zuber J., Fellmann C., McJunkin K., Miething C., Park Y., Dickins R.A., Hannon G.J., Lowe S.W.. A pipeline for the generation of shRNA transgenic mice. Nat. Protoc. 2012; 7:374–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vidigal J.A., Morkel M., Wittler L., Brouwer-Lehmitz A., Grote P., Macura K., Herrmann B.G.. An inducible RNA interference system for the functional dissection of mouse embryogenesis. Nucleic Acids Res. 2010; 38:e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tumbar T., Guasch G., Greco V., Blanpain C., Lowry W.E., Rendl M., Fuchs E.. Defining the epithelial stem cell niche in skin. Science (New York, N.Y.). 2004; 303:359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cedar H., Bergman Y.. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009; 10:295–304. [DOI] [PubMed] [Google Scholar]
- 22. Schubeler D., Lorincz M.C., Cimbora D.M., Telling A., Feng Y.Q., Bouhassira E.E., Groudine M.. Genomic targeting of methylated DNA: influence of methylation on transcription, replication, chromatin structure, and histone acetylation. Mol. Cell. Biol. 2000; 20:9103–9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schubeler D. Function and information content of DNA methylation. Nature. 2015; 517:321–326. [DOI] [PubMed] [Google Scholar]
- 24. Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L. et al. . Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science (New York, N.Y.). 2009; 324:930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu X., Zhao B.S., He C.. TET family proteins: oxidation activity, interacting molecules, and functions in diseases. Chem. Rev. 2015; 115:2225–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rasmussen K.D., Helin K.. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016; 30:733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu H., Zhang Y.. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011; 25:2436–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang T., Cooper S., Brockdorff N.. The interplay of histone modifications—writers that read. EMBO Rep. 2015; 16:1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kues W.A., Schwinzer R., Wirth D., Verhoeyen E., Lemme E., Herrmann D., Barg-Kues B., Hauser H., Wonigeit K., Niemann H.. Epigenetic silencing and tissue independent expression of a novel tetracycline inducible system in double-transgenic pigs. FASEB J. 2006; 20:1200–1202. [DOI] [PubMed] [Google Scholar]
- 30. Oyer J.A., Chu A., Brar S., Turker M.S.. Aberrant epigenetic silencing is triggered by a transient reduction in gene expression. PLoS ONE. 2009; 4:e4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pankiewicz R., Karlen Y., Imhof M.O., Mermod N.. Reversal of the silencing of tetracycline-controlled genes requires the coordinate action of distinctly acting transcription factors. J. Gene Med. 2005; 7:117–132. [DOI] [PubMed] [Google Scholar]
- 32. Zambrowicz B.P., Imamoto A., Fiering S., Herzenberg L.A., Kerr W.G., Soriano P.. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:3789–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Giel-Moloney M., Krause D.S., Chen G., Van Etten R.A., Leiter A.B.. Ubiquitous and uniform in vivo fluorescence in ROSA26-EGFP BAC transgenic mice. Genesis (New York, N.Y.). 2007; 45:83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haenebalcke L., Goossens S., Naessens M., Kruse N., Farhang Ghahremani M., Bartunkova S., Haigh K., Pieters T., Dierickx P., Drogat B. et al. . Efficient ROSA26-based conditional and/or inducible transgenesis using RMCE-compatible F1 hybrid mouse embryonic stem cells. Stem Cell Rev. 2013; 9:774–785. [DOI] [PubMed] [Google Scholar]
- 35. Papapetrou E.P., Lee G., Malani N., Setty M., Riviere I., Tirunagari L.M., Kadota K., Roth S.L., Giardina P., Viale A. et al. . Genomic safe harbors permit high β-globin transgene expression in thalassemia induced pluripotent stem cells. Nat. Biotechnol. 2011; 29:73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schucht R., Coroadinha A.S., Zanta-Boussif M.A., Verhoeyen E., Carrondo M.J., Hauser H., Wirth D.. A new generation of retroviral producer cells: predictable and stable virus production by Flp-mediated site-specific integration of retroviral vectors. Mol. Ther. 2006; 14:285–292. [DOI] [PubMed] [Google Scholar]
- 37. Sandhu U., Cebula M., Behme S., Riemer P., Wodarczyk C., Metzger D., Reimann J., Schirmbeck R., Hauser H., Wirth D.. Strict control of transgene expression in a mouse model for sensitive biological applications based on RMCE compatible ES cells. Nucleic Acids Res. 2011; 39:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rand U., Riedel J., Hillebrand U., Shin D., Willenberg S., Behme S., Klawonn F., Koster M., Hauser H., Wirth D.. Single-cell analysis reveals heterogeneity in onset of transgene expression from synthetic tetracycline-dependent promoters. Biotechnol. J. 2015; 10:323–331. [DOI] [PubMed] [Google Scholar]
- 39. Urlinger S., Baron U., Thellmann M., Hasan M.T., Bujard H., Hillen W.. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:7963–7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schaft J., Ashery-Padan R., van der Hoeven F., Gruss P., Stewart A.F.. Efficient FLP recombination in mouse ES cells and oocytes. Genesis. 2001; 31:6–10. [DOI] [PubMed] [Google Scholar]
- 41. Suda T., Liu D.. Hydrodynamic gene delivery: its principles and applications. Mol. Ther. 2007; 15:2063–2069. [DOI] [PubMed] [Google Scholar]
- 42. Ehrich M., Nelson M.R., Stanssens P., Zabeau M., Liloglou T., Xinarianos G., Cantor C.R., Field J.K., van den Boom D.. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15785–15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klug M., Heinz S., Gebhard C., Schwarzfischer L., Krause S.W., Andreesen R., Rehli M.. Active DNA demethylation in human postmitotic cells correlates with activating histone modifications, but not transcription levels. Genome Biol. 2010; 11:R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Blaschke K., Ebata K.T., Karimi M.M., Zepeda-Martinez J.A., Goyal P., Mahapatra S., Tam A., Laird D.J., Hirst M., Rao A. et al. . Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013; 500:222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Young J.I., Zuchner S., Wang G.. Regulation of the epigenome by Vitamin C. Annu. Rev. Nutr. 2015; 35:545–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Al-Dosari M., Zhang G., Knapp J.E., Liu D.. Evaluation of viral and mammalian promoters for driving transgene expression in mouse liver. Biochem. Biophys. Res. Commun. 2006; 339:673–678. [DOI] [PubMed] [Google Scholar]
- 47. Akhtar W., de Jong J., Pindyurin A.V., Pagie L., Meuleman W., de Ridder J., Berns A., Wessels L.F., van Lohuizen M., van Steensel B.. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013; 154:914–927. [DOI] [PubMed] [Google Scholar]
- 48. Kruse N., Spencer S., Wirth D.. Hauser H, Wagner R. Animal cell biotechnology in biologics production. 2014; De Gruyter; 173–216. [Google Scholar]
- 49. Nehlsen K., Schucht R., da Gama-Norton L., Kromer W., Baer A., Cayli A., Hauser H., Wirth D.. Recombinant protein expression by targeting pre-selected chromosomal loci. BMC Biotechnol. 2009; 9:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nehlsen K., da Gama-Norton L., Schucht R., Hauser H., Wirth D.. Towards rational engineering of cells: Recombinant gene expression in defined chromosomal loci. BMC Proc. 2011; 5(Suppl. 8):O6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gaj T., Gersbach C.A., Barbas C.F. 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013; 31:397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tchorz J.S., Suply T., Ksiazek I., Giachino C., Cloetta D., Danzer C.P., Doll T., Isken A., Lemaistre M., Taylor V. et al. . A modified RMCE-compatible Rosa26 locus for the expression of transgenes from exogenous promoters. PLoS One. 2012; 7:e30011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Strathdee D., Ibbotson H., Grant S.G.. Expression of transgenes targeted to the Gt(ROSA)26Sor locus is orientation dependent. PLoS ONE. 2006; 1:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wortge S., Eshkind L., Cabezas-Wallscheid N., Lakaye B., Kim J., Heck R., Abassi Y., Diken M., Sprengel R., Bockamp E.. Tetracycline-controlled transgene activation using the ROSA26-iM2-GFP knock-in mouse strain permits GFP monitoring of DOX-regulated transgene-expression. BMC Dev. Biol. 2010; 10:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Backman C.M., Zhang Y., Malik N., Shan L., Hoffer B.J., Westphal H., Tomac A.C.. Generalized tetracycline induced Cre recombinase expression through the ROSA26 locus of recombinant mice. J. Neurosci. Methods. 2009; 176:16–23. [DOI] [PubMed] [Google Scholar]
- 56. Wan M., Gu H., Wang J., Huang H., Zhao J., Kaundal R.K., Yu M., Kushwaha R., Chaiyachati B.H., Deerhake E. et al. . Inducible mouse models illuminate parameters influencing epigenetic inheritance. Development. 2013; 140:843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wolf D., Goff S.P.. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009; 458:1201–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jahner D., Jaenisch R.. Retrovirus-induced de novo methylation of flanking host sequences correlates with gene inactivity. Nature. 1985; 315:594–597. [DOI] [PubMed] [Google Scholar]
- 59. Rebollo R., Miceli-Royer K., Zhang Y., Farivar S., Gagnier L., Mager D.L.. Epigenetic interplay between mouse endogenous retroviruses and host genes. Genome Biol. 2012; 13:R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Turker M.S. Gene silencing in mammalian cells and the spread of DNA methylation. Oncogene. 2002; 21:5388–5393. [DOI] [PubMed] [Google Scholar]
- 61. Zeng H., Horie K., Madisen L., Pavlova M.N., Gragerova G., Rohde A.D., Schimpf B.A., Liang Y., Ojala E., Kramer F. et al. . An inducible and reversible mouse genetic rescue system. PLoS Genet. 2008; 4:e1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Maeder M.L., Angstman J.F., Richardson M.E., Linder S.J., Cascio V.M., Tsai S.Q., Ho Q.H., Sander J.D., Reyon D., Bernstein B.E. et al. . Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat. Biotechnol. 2013; 31:1137–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xu X., Tao Y., Gao X., Zhang L., Li X., Zou W., Ruan K., Wang F., Xu G.L., Hu R.. A CRISPR-based approach for targeted DNA demethylation. Cell Discov. 2016; 2:16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Morita S., Noguchi H., Horii T., Nakabayashi K., Kimura M., Okamura K., Sakai A., Nakashima H., Hata K., Nakashima K. et al. . Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat. Biotechnol. 2016; 34:1060–1065. [DOI] [PubMed] [Google Scholar]
- 65. Liu X.S., Wu H., Ji X., Stelzer Y., Wu X., Czauderna S., Shu J., Dadon D., Young R.A., Jaenisch R.. Editing DNA methylation in the mammalian genome. Cell. 2016; 167:233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stelzer Y., Shivalila C.S., Soldner F., Markoulaki S., Jaenisch R.. Tracing dynamic changes of DNA methylation at single-cell resolution. Cell. 2015; 163:218–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Amabile A., Migliara A., Capasso P., Biffi M., Cittaro D., Naldini L., Lombardo A.. Inheritable silencing of endogenous genes by hit-and-run targeted epigenetic editing. Cell. 2016; 167:219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rand U., Hillebrand U., Sievers S., Willenberg S., Koster M., Hauser H., Wirth D.. Uncoupling of the dynamics of host-pathogen interaction uncovers new mechanisms of viral interferon antagonism at the single-cell level. Nucleic Acids Res. 2014; 42:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bintu L., Yong J., Antebi Y.E., McCue K., Kazuki Y., Uno N., Oshimura M., Elowitz M.B.. Dynamics of epigenetic regulation at the single-cell level. Science (New York, N.Y.). 2016; 351:720–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Beard C., Hochedlinger K., Plath K., Wutz A., Jaenisch R.. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. 2006; 44:23–28. [DOI] [PubMed] [Google Scholar]
- 71. Wiznerowicz M., Jakobsson J., Szulc J., Liao S., Quazzola A., Beermann F., Aebischer P., Trono D.. The Kruppel-associated box repressor domain can trigger de novo promoter methylation during mouse early embryogenesis. J. Biol. Chem. 2007; 282:34535–34541. [DOI] [PubMed] [Google Scholar]
- 72. Baron U., Freundlieb S., Gossen M., Bujard H.. Co-regulation of two gene activities by tetracycline via a bidirectional promoter. Nucleic Acids Res. 1995; 23:3605–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.