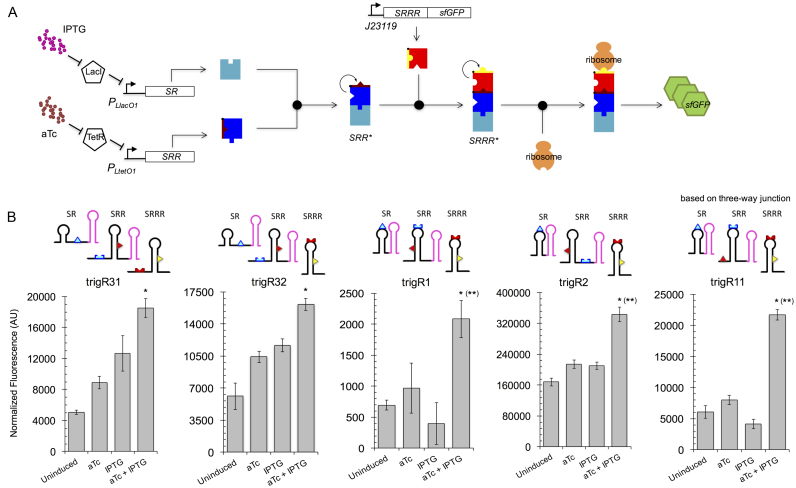
Figure 3.
Functional characterization of designer RNA hybridization networks in bacterial cell populations. (A) Scheme of the engineered sRNA circuit. Promoters PLlacO1 and PLtetO1 control the expression of the two sRNAs (SR and SRR), which can be tuned with different concentrations of the external inducers IPTG and aTc, whereas the mRNA (SRRR:sfGFP) is constitutively expressed from promoter J23119. The two sRNAs first interact to form a complex that is then able to activate a cis-repressed gene. The reporter gene encodes a sfGFP. (B) Fluorescence results (arbitrary units, AU) from the sRNA systems trigR31, trigR32, trigR1, trigR2 and trigR11 for all possible combinations of inducers. Error bars represent standard deviations over three biological replicates. The structural schemes of each single species implementing a system are shown. In each case, the asterisk (or two asterisks in brackets) denotes P < 0.05, one-tailed Welch t-test, comparing the fluorescence level when both inducers are present with respect to the level when there is only one inducer (or the level reached by the additive effect of the two inducers).