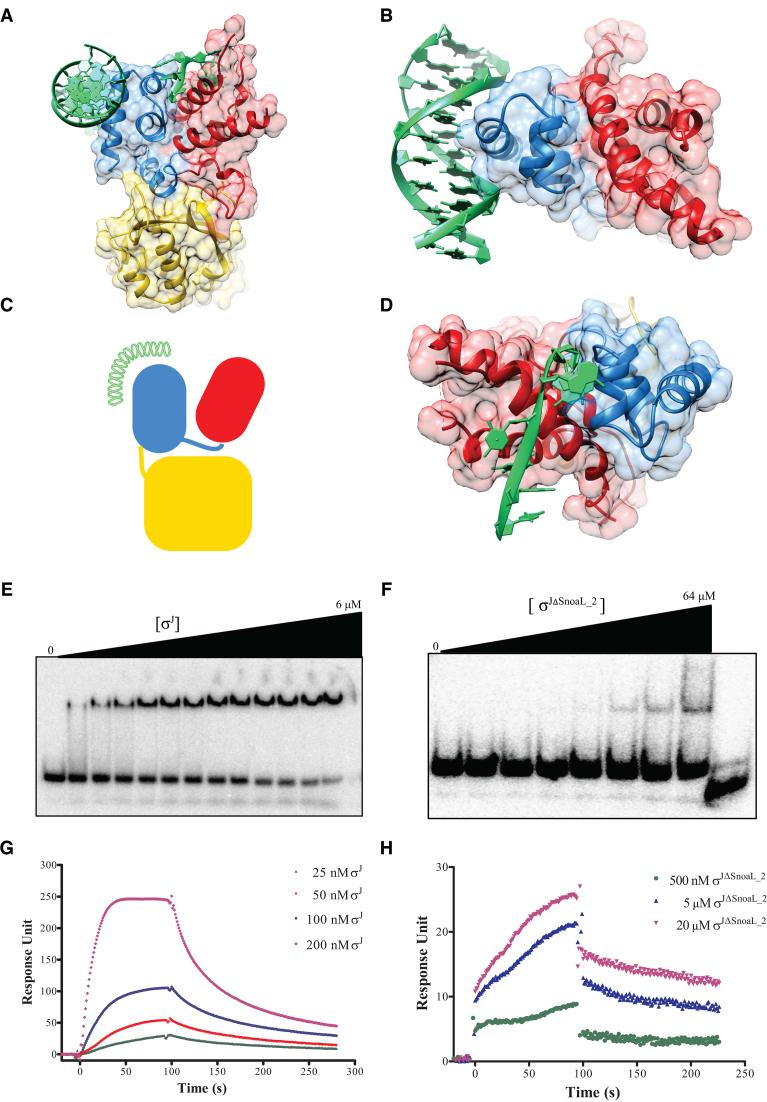
Figure 2.
Exposed DNA binding surfaces suggest that σJ adopts a conformation that can readily interact with the promoter. (A and B) Mapping of the −10 and −35 promoter DNA on σJ2 and σJ4 by superposition with previously determined structures of the −10 promoter/σ2 (PDB ID: 4LUP) and −35 promoter/σ4 (PDB ID: 2H27) complexes (60,63). (C) A schematic representation of DNA binding to σJ. (D) The exposed DNA binding surfaces of σJ2 and σJ4 reveals that the Snoal_2 domain tethers the two DNA binding domains in this conformation but is unlikely to interact with the DNA. (E) Radiolabeled sigI-promoter DNA was incubated with (E) σJ (0–6 μM) or (F) σJΔSnoaL_2 (0–64 μM) and separated on a non-denaturing PAGE. DNA binding is significantly reduced in σJΔSnoaL_2. The wedge at the top of each panel depicts the gradient of protein concentration. The concentrations of σJ from left to right are 0, 1, 1.5, 1.75, 2, 2.25, 2.5, 2.75, 3, 3.5, 4, 5 and 6 μM respectively. Similarly, the concentration of σJΔSnoaL_2 from left to right are 0, 1, 2, 4, 8, 16, 32 and 64 μM respectively. The right-most lane in panel (F) is loaded with 5 μM σJ as a binding control. (G) In surface plasmon resonance experiments varying concentrations of (G) σJ and (H) σJΔSnoaL_2 were passed on a chip with an immobilized sigI-promoter. Put together, these experiments reveal that deletion of SnoaL_2 significantly impairs σJ–promoter DNA interaction.