Fig 8. Sly-MIR6020 generates 22-nt miRNA and triggers phasiRNA production.
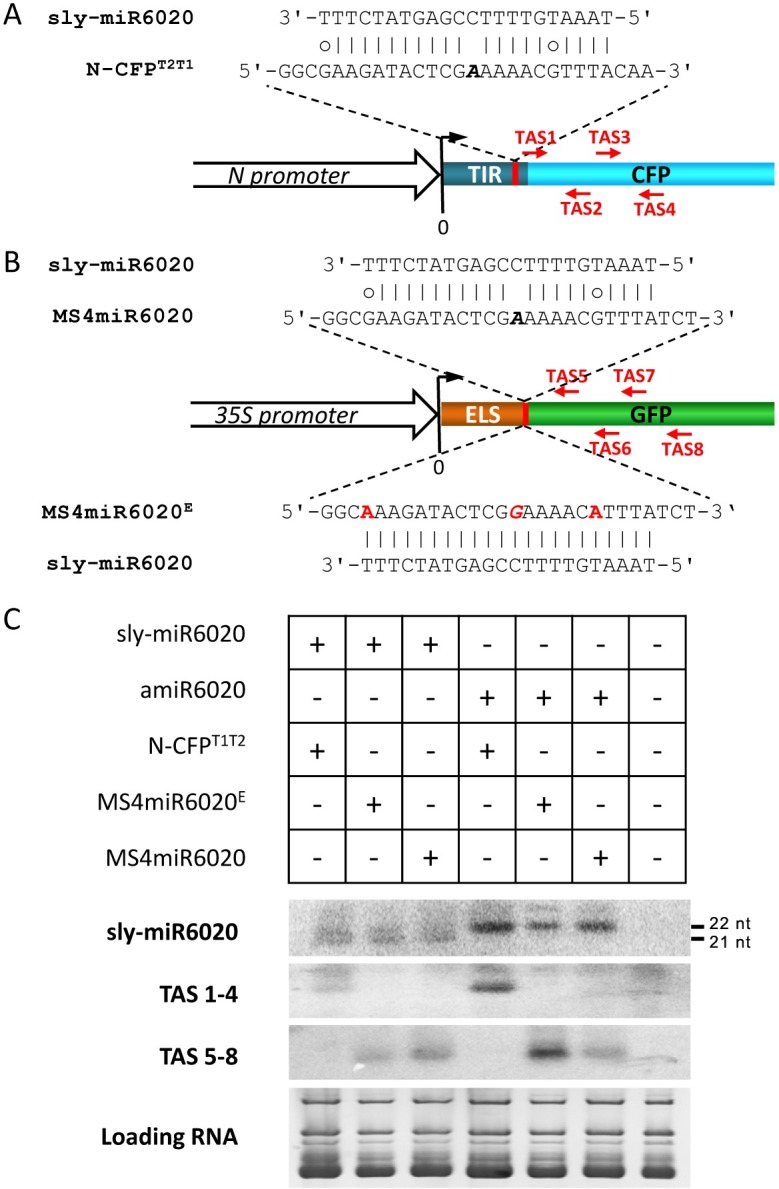
(A) Map of sly-miR6020::N-CFPT2T1 sequence alignment and four CFP-phasiRNA (red arrows) down-stream of the miR6020 binding site. The sequence of sly-miR6020 binding site (red area) is shown above the N-CFPT2T1 construct map. Bases in italics indicate the cleavage site. (B) A map of sly-miR6020::MS4miR6020 sequence alignment and four GFP-phasiRNA (red arrows) down-stream of the miR6020 binding site. The sequences of sly-miR6020 binding site (red area) in MS4miR6020 and MS4miR6020E are shown above and below the construct map respectively. Bases in italics indicate the cleavage site. The mutated bases are indicated in red. ELS, Endoplasmic reticulum (ER) localization signal. (C) Northern blot detection of phasiRNAs triggered by sly-miR6020. Probes are indicated to the left. EB staining of tRNA and rRNA serves as a loading control.
