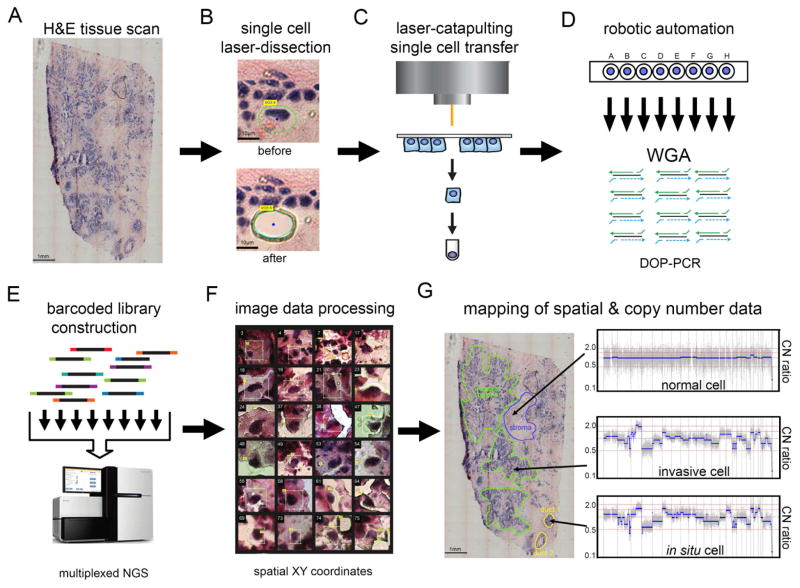
Figure 1. Topographic Single Cell Sequencing of DCIS Tissues.
(A) Whole-tissue scanning is performed on H&E stained synchronous DCIS tissues at low 10X magnification. (B) UV laser-microdissection of a single cell at 63X magnification (C) laser-catapulting transfer of a single cell into a collection tube. (D) automated robotic depositing of single cells into 8-well strip tubes with lysis buffer into a 96-well manifold, followed by whole-genome-amplification using DOP-PCR. (E) Construction of barcoded single cell libraries for multiplexed pooling and sparse whole-genome sequencing on the Illumina platform. (F) Processing of brightfield images of single cells and spatial XY coordinates. (G) Mapping of spatial coordinates and genomic data in tissue sections, showing examples of genomic copy number profiles from a normal cell, in situ tumor cell, and an invasive tumor cell.