Fig. 3.
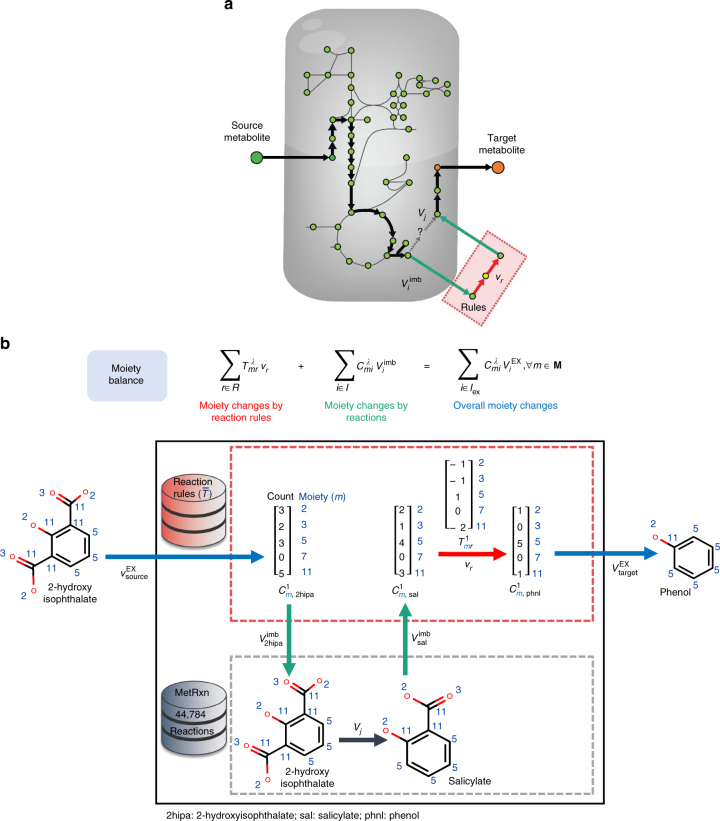
The novoStoic procedure. novoStoic seamlessly blend existing reactions and reaction rules while designing a pathway from source metabolite to target metabolite. a The variable defines the connection (green arrows) between the known reactions network (black arrows) and the reaction rules network (red arrows and red dotted box). It indicates the surplus or deficit of metabolite i in the known metabolic network. The reaction rules network is invoked to complete a pathway when no known reaction can catalyze the conversion. b Illustration of the component balance and moiety balance defined in novoStoic constraints (2) and (3) (Supplementary Methods). The bottom half (gray dotted box) is the known network operating on metabolites, while the upper half (red dotted box) is the reaction rules network operating on moieties. The section of the entire system that is involved in the moiety balance constraint is color-coded: (red) moiety changes by reaction rules, (blue) overall moiety changes, and (green) moiety changes by known reactions. The pathway here converts 2hipa to phnl using a known reaction (i.e., 2hipa → sal, black arrow) and a reaction rule (i.e., sal → phnl, red arrow). Metabolite 2hipa first enters the reaction rule network using exchange reaction (blue arrow, ) and subsequently exported to the known reaction network (green arrow, ). It is converted by a known reaction (black arrow) into sal, which is then transferred to the reaction rule network (green arrow, ). The reaction rule (red arrow) converts sal into phnl, which is exported out of the entire system . Note that for clarity purpose, the involvement of carbon dioxide in the conversion of 2hipa to sal and sal to phnl is omitted