Abstract
As the cheapest energy source, carbohydrates are used in fish feeds to improve physical quality and reduce catabolism of proteins and lipids. The liver is the primary organ for metabolism and is also an important site of immune regulation. Here, we investigated the effect of different dietary carbohydrate levels on growth and health by evaluating the liver transcriptome of Epinephelus akaara. In this study, E. akaara juveniles were fed diets containing few (0% corn starch), moderate (18% corn starch), and high (30% corn starch) levels of dietary carbohydrate. After an 8-week feeding trial, E. akaara fed 30% dietary carbohydrates exhibited poor growth performance compared with those fed 0% and 18% dietary carbohydrates (P > 0.05). Genes related to the immune system, including IL8, TLR9, CXCR4, CCL4, and NFκB inhibitor alpha, were over-expressed in E. akaara fed the highest level of carbohydrate (30%). This general over-expression could indicate activation of inflammatory processes in the liver. The liver transcriptome data of E. akaara reported here indicate that high carbohydrate level of diet can lead to poor growth and inflammatory immune response in E. akaara.
Introduction
Carbohydrates are the cheapest energy sources and the major compounds that make up organisms. Some studies have indicated that certain levels of carbohydrates can improve feed utilization and protein retention in rainbow trout (Oncorhynchus mykiss), Atlantic salmon (Salmo salar), European eel (Anguilla anguilla), Atlantic cod (Gadus morhua), and different carp species (Catla catla, Labeo rohita, Cirrhinus mrigala)1–7. If sufficient carbohydrate content is not provided in the diet, other nutrients such as proteins and lipid can be used for energy8; This could lead to an increase in cost and nutritional imbalances. In addition, post-prandial prolonged hyperglycemia occurs when a high level of carbohydrates is consumed9–14. An imbalance carbohydrates in the diet may make the fish under metabolic stress15 and have negative effects on nutrient retention, growth, metabolism, and health16. Thus, it is important to supply an appropriate level of carbohydrates in aqua-feeds.
The liver is the primary target organ for metabolism. Nutrients are absorbed into the body through the small intestine and then delivered to liver. The critical metabolic functions of the liver often eclipse its role as an important organ for immune regulation17, as the liver serves as a physical barrier responsible for filtering potentially harmful antigens, which may reach the body via the gastrointestinal tract. Castro et al.18 reported that the liver modulates the immune response following hemorrhagic septicemia virus (VHSV) infection in rainbow trout. Similarly, when inappropriate feed supplied, the liver also can function as an immunocompetent organ19–22. A growing body of research has focused on the relationship between carbohydrate supplementation and immune function. For example, carbohydrates can regulate the production of proinflammatory cytokines to enhance the endurance performance and attenuated stress hormone response23.
Carbohydrate utilization differs among fish species16,24–26. Herbivorous and omnivorous fish can utilize as much as 45% carbohydrate content in diet27–29, but carnivorous fish show significantly poorer growth when fed diets with 30% carbohydrate levels compared to those fed moderate carbohydrate levels (in general, ≤20%), such as Atlantic salmon5 and cobia (Rachycentron canadum)30. Epinephelus akaara (Temminck and Schlegel, 1842) is an important marine carnivorous fish with a high market value in Asia, To date, few nutrient requirements have been investigated in E. akaara, except for appropriate level of proteins31 and lipids32. To obtain the appropriate formulation of fish feed, it is an essential pre-requisite to understand the capacity of fish to utilize carbohydrates8. High-throughput sequencing can provide an unprecedented view of global gene expression and detailed molecular information on responses to nutrition metabolism. Therefore, we here focused on the liver transcriptome of E. akaara fed diets containing few (0%), moderate (18%), and high (30%) levels of carbohydrates using high-throughput sequencing to identify genes responsible for growth and immune system alterations.
Results
Growth performance and growth-related gene expression in the liver
The growth performance of E. akaara was measured by percentage weight gain (PWG). In this study, the final body weight (g/fish) in each group was 29.30 ± 3.32, 28.54 ± 2.87, and 24.02 ± 0.92, respectively; the PWG (%) were 276.16 ± 41.99, 266.26 ± 28.33, and 208.69 ± 12.68 in the C1, C2, and C3 groups, respectively. PWG generally decreased with increasing dietary carbohydrate (P > 0.05), and fish fed 30% carbohydrates exhibited the lowest PWG.
We evaluated genes belonging to the GH/IGF-system to show the relationship between growth and gene expression. Several components of the GH/IGF axis in each group exhibited different expression levels. The expression of GHR, IGF1, IGF2, IGFBP2, IGFBP3 (P < 0.05), IGFBP4, and IGFBP6 (P > 0.05) were decreased in the C2 and C3 groups compared to the C1 group, whereas IGFBP1 (P < 0.05), IGFBP5, and IGFBP7 (P > 0.05) were upregulated in the C3 group to levels comparable to the C1 and C2 groups (Table 1).
Table 1.
The expression of growth-related genes in three groups.
| Gene ID | name | C1 | C2 | C3 |
|---|---|---|---|---|
| c56634_g3 | GHR | 11.8233 | 8.4724 | 6.9467 |
| c49944_g2 | IGFBP1 | 1.6992 | 4.3513 | 84.0755 |
| c46871_g1 | IGFBP2A | 12.4657 | 11.8959 | 5.4010 |
| c59085_g2 | IGFBP2B | 102.9990 | 97.3850 | 65.5878 |
| c33486_g1 | IGFBP4 | 0.9967 | 0.7571 | 0.7533 |
| c44323_g3 | IGFBP5 | 16.8376 | 20.6902 | 24.5923 |
| c69225_g1 | IGFBP6 | 0.6224 | 0.3081 | 0.1927 |
| c49986_g1 | IGFBP7 | 2.6987 | 3.8087 | 4.8402 |
| c60956_g7 | IGF1 | 78.6293 | 29.1721 | 17.9944 |
| c59327_g1 | IGF2 | 49.4954 | 6.9903 | 10.1116 |
De novo assembly
There were 5,5912,920, 63,532,872, and 78,789,902 raw reads in the C1, C2, and C3 groups, respectively, generated by high-throughput sequencing of the cDNA library of the E. akaara liver. We cleaned the low-quality reads; the sequence of high quality rates were 87.13% (clean reads number: 48,717,774), 88.10% (55,971,730), 85.33% (67,230,310) in C1, C2, and C3 groups, respectively. There were 94% sequences up to the quality score of Q30, which describes quality score logarithmically linked to error probabilities (i.e., Q30 = 99.9%, chance correct base called). The sequencing results showed that these data were appropriate for analysis (Table 2). The ORF predictions came from the contigs assembled using Trinity. The contigs in each group were 83,451, 84,162, and 94,733, respectively. Subsequently, to annotate the sequences, blastp and blastx alignments (E-value < 10−5) with the NT, NR, gene, and string were used, and 27,327, 27,851, 31,234 contigs in each group featured a corresponding annotation (Table 2).
Table 2.
Sequencing information of the E. akaara liver.
| Samples | C1 | C2 | C3 |
|---|---|---|---|
| Raw Reads Number | 55,912,920 | 63,532,872 | 78,789,902 |
| Clean Reads Number | 48,717,774 | 55,971,730 | 67,230,310 |
| Clean Reads Rate(%) | 87.13 | 88.10 | 85.33 |
| Low-quality Reads Number | 4,827,778 | 5,897,502 | 7,384,488 |
| Low-quality Reads Rate(%) | 8.63 | 9.28 | 9.37 |
| Ns Reads Number | 3,910 | 4,422 | 5,330 |
| Ns Reads Rate(%) | 0.01 | 0.01 | 0.01 |
| Adapter Polluted Reads Number | 2,363,458 | 1,659,218 | 4,169,774 |
| Adapter Polluted Reads Rate(%) | 4.23 | 2.61 | 5.29 |
| Raw Q30 Bases Rate(%) | 89.93 | 90.04 | 88.98 |
| Clean Q30 Bases Rate(%) | 94.84 | 94.86 | 94.67 |
| ORF counts | 27327 | 27851 | 31234 |
| ORF N50 (bp) | 1011 | 1098 | 1074 |
Functional annotation and analysis of differentially-expressed genes
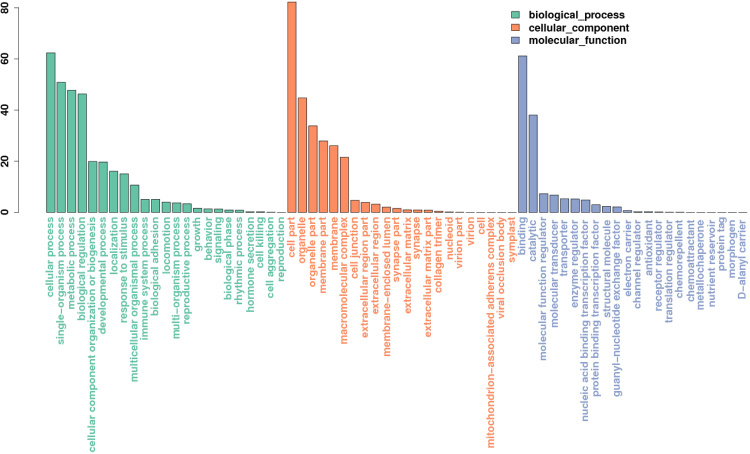
GO terms for the transcriptome were analyzed using Blast2GO, which provides information on “Biological Processes”, “Cellular Components” and “Molecular Function” for each contig (Fig. 1). In the “biological processes” categories, which features 23 subtypes, most corresponding genes were involved in cellular processes, single-organism processes, metabolic processes, and biological regulation. In addition, 22 subtypes were annotated with “cellular components”; most corresponding genes were involved in the cell, organelles, and parts of the membrane. In the “molecular function” category, which featured 22 subtypes, most corresponding genes were involved with binding and catalytic activity.
Figure 1.
GO classification of assembled genes in the E. akaara liver transcriptome.
The KEGG database was used to obtain more information to predict the unigene functions; 38,938 genes ware classified into 113 KEGG pathways. The KEGG pathway analysis was also used to identify genes observed to be differentially expressed in the C1-C2, C2-C3, and C1-C3 pair groups fed few (C1), moderate (C2), or high (C3) levels of carbohydrates. The numbers of differentially-expressed genes in each group (C1-C2, C2-C3, and C1-C3) were 20,499, 35,984, and 37,203, respectively (Fig. 2). There were 20,499 genes differentially expressed in the C2 group relative to the C1 group, 35,984 genes differently expressed in the C3 group relative to the C2 group, and 37,203 genes differently expressed in the C3 group relative to the C2 group. Compared with C1 and C2 group, the number of up-regulated gene in C3 group is no significant difference (P > 0.05).
Figure 2.
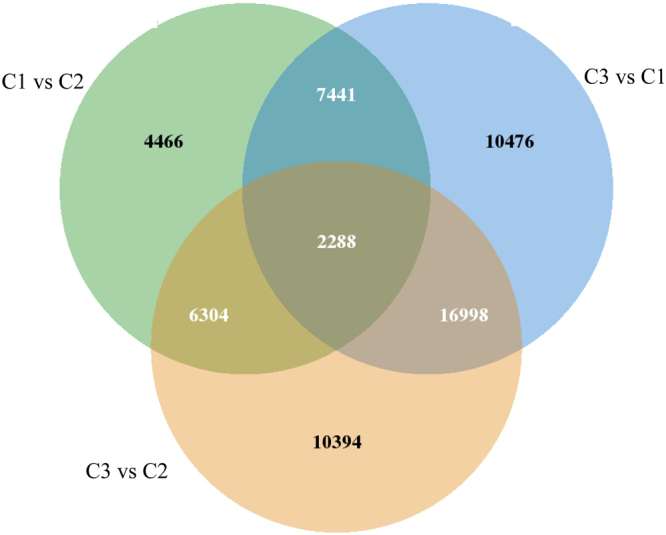
Differentially-expressed gene distribution. For example, 16,998 genes are differentially-expressed in C3 group compared with C1 and C2 groups.
We also chose categories including “Carbohydrate Metabolism”, “Energy Metabolism”, “Lipid Metabolism”, “Amino Acid Metabolism”, ‘Metabolism of Other Amino Acids”, “Glycan Biosynthesis and Metabolism”, “Immune System” and “Digestive System” (Table 3) to evaluate metabolic stress information among the three groups. There were 15 pathways related to the immune system, and most genes were over-expressed in the C3 group compared to the C1 and C2 groups (Table 3). In the chemokine signaling pathway, there were 153 and 144 genes up-regulated in the C3 group compared with the C1 and C2 groups. In addition, we chose 20 genes which belonged to immune system to further confirm the results of differentially-expressed analysis using RT-qPCR (Fig. 3).
Table 3.
The number of genes whose expression changed in the metabolism pathway between two groups.
| Pathway | c1-c2 up | c1-c2 down | c1-c3 up | c1-c3 down | c3-c2 up | c3-c2 down | |
|---|---|---|---|---|---|---|---|
| Carbohydrate metabolism | |||||||
| 00562 | Inositol phosphate metabolism | 3 | 29 | 45 | 11 | 53 | 6 |
| 00010 | Glycolysis/Gluconeogenesis | 17 | 8 | 52 | 7 | 32 | 8 |
| 00500 | Starch and sucrose metabolism | 10 | 6 | 17 | 19 | 18 | 19 |
| 00520 | Amino sugar and nucleotide sugar metabolism | 15 | 2 | 31 | 4 | 18 | 11 |
| 00620 | Pyruvate metabolism | 25 | 3 | 25 | 3 | 18 | 15 |
| 00051 | Fructose and mannose metabolism | 17 | 3 | 37 | 3 | 21 | 8 |
| 00052 | Galactose metabolism | 11 | 4 | 29 | 7 | 18 | 7 |
| 00640 | Propanoate metabolism | 19 | 0 | 4 | 1 | 3 | 13 |
| 00020 | Citrate cycle(TCA cycle) | 6 | 4 | 16 | 3 | 13 | 2 |
| 00040 | Pentose and glucuronate interconversions | 1 | 1 | 4 | 8 | 7 | 6 |
| 00030 | Pentose phosphate pathway | 8 | 7 | 20 | 7 | 11 | 5 |
| 00053 | Ascorbate and aldarate metabolism | 2 | 2 | 0 | 8 | 4 | 6 |
| 00630 | Glyoxylate and dicarboxylate metabolism | 0 | 2 | 0 | 2 | 2 | 2 |
| 00650 | Butanoate metabolism | 2 | 2 | 4 | 1 | 4 | 2 |
| Energy metabolism | |||||||
| 00190 | Oxidative phosphorylation | 4 | 19 | 25 | 4 | 46 | 0 |
| 00680 | Methane metabolism | 14 | 8 | 22 | 7 | 9 | 5 |
| 00710 | Carbon fixation in photosynthetic organisms | 1 | 5 | 10 | 7 | 8 | 4 |
| 00720 | Carbon fixation pathways in prokaryotes | 2 | 0 | 3 | 0 | 2 | 1 |
| 00195 | Photosynthesis | 0 | 0 | 0 | 0 | 1 | 0 |
| 00920 | Sulfur metabolism | 2 | 0 | 3 | 0 | 2 | 0 |
| 00910 | Nitrogen metabolism | 4 | 1 | 2 | 0 | 2 | 3 |
| Lipid metabolism | |||||||
| 00564 | Glycerophospholipid metabolism | 15 | 23 | 35 | 32 | 33 | 15 |
| 00561 | Glycerolipid metabolism | 13 | 17 | 25 | 26 | 23 | 12 |
| 00600 | Sphingolipid metabolism | 7 | 2 | 13 | 6 | 14 | 2 |
| 00071 | Fatty acid degradation | 1 | 3 | 6 | 8 | 7 | 9 |
| 00590 | Arachidonic acid metabolism | 7 | 6 | 17 | 24 | 17 | 12 |
| 00565 | Ether lipid metabolism | 5 | 4 | 17 | 9 | 16 | 3 |
| 00140 | Steroid hormone biosynthesis | 4 | 11 | 6 | 18 | 5 | 12 |
| 0062 | Fatty acid elongation | 1 | 5 | 12 | 9 | 12 | 5 |
| 00100 | Steroid biosynthesis | 14 | 25 | 16 | 8 | 23 | 4 |
| 01040 | Biosynthesis of unsaturated fatty acids | 1 | 9 | 5 | 10 | 6 | 3 |
| 00591 | Linoleic acid metabolism | 2 | 4 | 8 | 18 | 6 | 7 |
| 00120 | Primary bile acid biosynthesis | 2 | 5 | 3 | 9 | 1 | 5 |
| 00592 | alpha-Linolenic acid metabolism | 0 | 5 | 6 | 9 | 8 | 3 |
| 00061 | Fatty acid biosynthesis | 17 | 0 | 1 | 0 | 0 | 1 |
| 00072 | Synthesis and degradation of ketone bodies | 0 | 1 | 0 | 0 | 3 | 0 |
| 00073 | Cutin, suberine and wax biosynthesis | 5 | 0 | 5 | 0 | 3 | 1 |
| Amino acid metabolism | |||||||
| 00310 | Lysine degradation | 7 | 20 | 19 | 18 | 23 | 11 |
| 00330 | Arginine and proline metabolism | 8 | 6 | 16 | 7 | 15 | 7 |
| 00270 | Cysteine and methionine metabolism | 0 | 7 | 11 | 12 | 10 | 9 |
| 00280 | Valine, leucine and isoleucine degradation | 0 | 1 | 3 | 2 | 7 | 2 |
| 00260 | Glycine, serine and threonine metabolism | 7 | 11 | 14 | 8 | 7 | 4 |
| 00380 | Tryptophan metabolism | 4 | 4 | 6 | 1 | 7 | 2 |
| 00250 | Alanine, aspartate and glutamate metabolism | 0 | 4 | 6 | 2 | 5 | 4 |
| 00340 | Histidine metabolism | 1 | 5 | 3 | 3 | 7 | 2 |
| 00350 | Tyrosine metabolism | 0 | 6 | 7 | 3 | 7 | 0 |
| 00360 | Phenylalanine metabolism | 1 | 5 | 6 | 3 | 7 | 1 |
| 00300 | Lysine biosynthesis | 0 | 1 | 0 | 2 | 0 | 0 |
| 00290 | Valine, leucine and isoleucine biosynthesis | 0 | 0 | 1 | 1 | 0 | 1 |
| Immune system | |||||||
| 04062 | Chemokine signaling pathway | 26 | 13 | 153 | 12 | 144 | 7 |
| 04670 | Leukocyte transendothelial migration | 25 | 16 | 120 | 18 | 110 | 10 |
| 04650 | Natural killer cell mediated cytotoxicity | 25 | 11 | 104 | 9 | 88 | 7 |
| 04660 | T cell receptor signaling pathway | 27 | 17 | 103 | 13 | 88 | 8 |
| 04666 | Fc gamma R-mediated phagocytosis | 18 | 19 | 101 | 14 | 92 | 9 |
| 04662 | B cell receptor signaling pathway | 23 | 7 | 93 | 10 | 77 | 8 |
| 04620 | Toll-like receptor signaling pathway | 17 | 10 | 76 | 8 | 61 | 7 |
| 04640 | Hematopoietic cell lineage | 23 | 4 | 67 | 7 | 48 | 12 |
| 04664 | Fc epsilon RI signaling pathway | 10 | 15 | 63 | 17 | 58 | 6 |
| 04621 | NOD-like receptor signaling pathway | 10 | 4 | 44 | 3 | 39 | 6 |
| 04622 | RIG-I-like receptor signaling pathway | 7 | 7 | 41 | 7 | 32 | 3 |
| 04672 | Intestinal immune network for IgA production | 6 | 0 | 25 | 0 | 18 | 0 |
| 04612 | Antigen processing and presentation | 3 | 3 | 23 | 6 | 22 | 7 |
| 04610 | Complement and coagulation cascades | 6 | 2 | 19 | 10 | 20 | 14 |
| 04623 | Cytosolic DNA-sensing pathway | 4 | 2 | 15 | 2 | 15 | 2 |
| Digestive system | |||||||
| 04972 | Pancreatic secretion | 11 | 25 | 65 | 4 | 63 | 6 |
| 04971 | Gastric acid secretion | 8 | 9 | 50 | 4 | 41 | 3 |
| 04970 | Salivary secretion | 9 | 11 | 42 | 6 | 38 | 5 |
| 04974 | Protein digestion and absorption | 14 | 20 | 95 | 5 | 91 | 5 |
| 04976 | Bile secretion | 9 | 14 | 31 | 16 | 28 | 9 |
| 04973 | Carbohydrate digestion and absorption | 5 | 4 | 25 | 9 | 27 | 7 |
| 04975 | Fat digestion and absorption | 8 | 8 | 8 | 25 | 4 | 16 |
| 04977 | Vitamin digestion and absorption | 3 | 6 | 1 | 25 | 1 | 6 |
| 04978 | Mineral absorption | 6 | 3 | 6 | 5 | 2 | 1 |
The data in table indicate genes involved in carbohydrate metabolism, energy metabolism, lipid metabolism, amino acid metabolism, immune system function, and digestive system function that were up-regulated and downregulated across groups.
Figure 3.
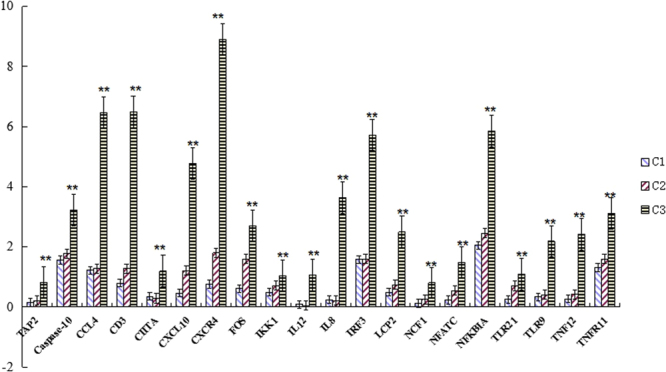
RT-qPCR confirmation of genes expressed at different carbohydrate level in C1, C2 and C3 groups. Bars represent mean ± standard error of three fish. Aasterisks indicate significant (P < 0.01) differences. The genes chosen for immune system were: APT2: Antigen peptide transporter 2; CCL4: C-C motif chemokine 4; CD3: T-cell surface glycoprotein CD3 delta chain; CIITA: MHC class II transactivator; CXCL10: C-X-C motif chemokine 10; CXCR4: C-X-C chemokine receptor type 4; FOS: Proto-oncogene c-Fos; IKK1: Inhibitor of nuclear factor kappa-B kinase subunit alpha; IL12: Interleukin-12 subunit beta; IL8: Interleukin-8; IRF3: Interferon regulatory factor 3; LCP2: Lymphocyte cytosolic protein 2; NCF1: Neutrophil cytosol factor 1; NFATC: Nuclear factor of activated T-cells; NFKBIA: NF-kappa-B inhibitor alpha; TLR21: Toll-like receptor 2 type-1; TLR9: Toll-like receptor 9; TNF12: Tumor necrosis factor 12; TNFR11: Tumor necrosis factor receptor superfamily member 11.
Discussion
As the cheapest energy sources, carbohydrates are used in fish feeds to improve physical quality and reduce catabolism of proteins and lipids33. However, carnivorous fish have a poor ability to utilize high level of carbohydrate. Thus, we investigated the effect of different dietary carbohydrate levels (0%, 18%, and 30%) on growth and health by evaluating the liver transcriptome in E. akaara.
In the present study, E. akaara fed a diet with 30% carbohydrate content exhibited poor growth performance (PWG = 208.69%) compared with those fed 0% (PWG = 276.16%) and 18% (PWG = 266.26%) carbohydrate content. These results indicated that PWG decreased with an increase in dietary carbohydrate levels. Various mechanisms could account for growth, including endocrine system change34. As in mammals, there are two major molecular targets, growth hormone (GH) and insulin-like growth factor (IGF), in the potential endocrine regulation of growth in the teleost35–38. They both belong to the GH/IGF-system, which also consists of multiple subtypes of GH receptors (GHRs) and insulin-like growth factor binding protein (IGFBP)39.
Several studies have confirmed that IGF and GHR gene expression which is closely related to growth performance can be modulated by the feed component40–45. Studies of coho salmon, gilthead sea bream (Sparus aurata), and tilapia have indicated that the levels of hepatic GHR and IGF mRNA are positively correlated with body growth rate46–50. In accord with these findings, E. akaara fed high levels of carbohydrates had the lowest PWG value and the lowest expression of IGF1 and GHR genes (Table 1).
In addition to IGF and GHR, IGFBPs are the main members of the GH/IGF-system. These IGFBPs are traditionally thought to function as carrier proteins and regulate circulating IGF turnover, transport, and distribution. And some studies have shown that IGFBPs are ubiquitously expressed across numerous tissues with autocrine or paracrine effects in salmonids and modulate IGF activities in target tissues44,51–53. Except for IGFBP-1, which is predominantly expressed in liver, all other IGFBPs are expressed in many peripheral tissues45. In our study, we found that the IGFBP 1, 5, 7 genes were up-regulated in fish fed high levels of carbohydrate diet (C3 group) compared to the other two groups. Elsewise, expression of the IGFBP 2, 3, 4, and 6 genes was reduced in the C3 group, The expression of multiple IGFBPs genes is regulated by different levels of carbohydrate, but little is known about their function in liver. And the mechanism is still not clear. These findings may reinforce the need for additional research about the relationship between IGFBPs gene expression and growth performance in fish.
The liver is an important organ for metabolism and immune function. Significant numbers of natural killer receptor-positive (NKR+) cells and macrophages reside in the healthy liver, which can detect foreign substances and produce inflammatory mediators. In the present study, most genes involved in innate and adaptive immune system were up-regulated following increased dietary carbohydrate levels, suggesting that high levels of carbohydrates can invoke an immune response.
There are many immune system pathways in organism, such as T-cell receptor signaling pathway, B cell receptor signaling pathway, chemokine signaling pathway, Toll-like receptor signaling pathway, RIG-I-like receptor signaling pathway, and NOD-like receptor signaling pathway. In our study, most genes which belongs to immune system pathways were up-regulated in C3 group which fed high carbohydrate level dietary compared with C1 and C2 groups which were fed with lower carbohydrate levels in diet (Table 3). We evaluated 20 genes using RT-qPCR, which belong to 15 pathways in the immune system, and found differential expression across the three groups (Fig. 3). Interleukin 8 (IL8) is a member of the CXC chemokine family and is one of the major mediators of the inflammatory response54,55, and, if the harmful antigens are present in the liver, it causes inflammatory-related genes to be over-expressed immediately. In our study, IL8 was more highly expressed in the group fed high levels of carbohydrates compared to the C1 and C2 groups, which had a low constitutive expression (Fig. 3). C-C motif chemokine 4 (CCL4), C-X-C motif chemokine 10 (CXCL10), and C-X-C chemokine receptor type 4 (CXCR4) genes also belong to the chemokine signaling pathway and are chemotactic factors which can active granulocytes, nature kill cells, and monocytes to product interleukin and interferon. In this study, these genes were significantly over-expressed in the group fed high levels of carbohydrates (Fig. 3), indicating the inflammation response in fish56.
In the T-cell receptor immune pathway, cluster of differentiation 3 (CD3) induces the T-cell receptor (TCR) to generate an activation signal in T lymphocytes57. This high specificity, combined with the presence of CD3 at all stages of T-cell development, makes it a useful immunohistochemical marker for T-cells in tissue sections. T- cells have largely diversified receptors to recognize different antigens, and the MHC class II plays a central role in determining T-cell-mediated adaptive immunity against various pathogens. In this study, MHC class II and CD3 genes had the highest expression in the group fed high levels of carbohydrates compared to the groups fed few and moderate levels. These results indicate that high levels of carbohydrates induced an adaptive immunity response.
Toll-like receptors (TLRs), inhibitor of nuclear factor kappa-B kinase subunit alpha (IKKA), caspase-8, proto-oncogene c-Fos (Fos), interferon regulatory factor 3 (IRF3), tumor necrosis factor (TNF), and tumor necrosis factor receptor (TNFR) belong to Toll-like receptor signaling pathway, NOD-like receptor signaling pathway, and RIG-I-like receptor signaling pathway. These three pathways are cytoplasmic pattern recognition receptors (PRRs), which are involved in the recognition of viruses by the innate immune system58. PRRs responded to many exogenous pathogens and/or endogenous serious signals, by recognizing highly conserved structures, such as pathogen associated molecular patterns (PAMPs) and danger/damage associated molecular patterns (DAMPs). Following recognition by PRRs, downstream signaling pathways induce expression of genes of inflammatory molecules that can activate the cytokine signaling pathway to regulate innate and adaptive immunity59. The genes belonging to these three pathways had significantly higher expression in the group fed high levels of carbohydrates, suggesting that an imbalance in carbohydrates signals the liver to produce immunocompetence through the innate and adaptive immune system.
In Atlantic salmon, another sea carnivorous fish, numerous studies reported plant feedstuffs could lead to the changes of histomorphological and immune response related genes expression in intestine at least four weeks60–65. However, Sahlmann et al.66 found that the gene expression changes could be detected in third day, which is earlier than signs of inflammation in histological evaluation. In this study, we got the liver transcriptome data of E. akaara after 8 weeks of rearing experiment and it showed that the fish consuming high level of carbohydrate may increase inflammation response. Then, we did a short feeding trial with C2 and C3 experimental diets to make up the defect of functional verification in original experiment. After 2 weeks, although there was no difference in histological section of liver tissue, the immune response related genes were higher expression in C3 group than C2 group by RT-qPCR. It suggested that the high level of carbohydrate had the tendency to give rise to immune reaction in E. akaara. On the other hand, we used 3 fishes of each replicating group per treatment as the RNA templet to confirmed the sequencing data with RT-qPCR. They were consistent with each other. These results all confirmed that our data and conclusion is credible.
Conclusion
The liver transcriptome data of E. akaara reported here indicate that high carbohydrate level of diet can lead to poor growth and inflammatory immune response in E. akaara. The up-regulation of a large number of genes in immune system pathway revealed the fish maybe experience inflammatory response which may be related with a decrease in growth rate. The prominent effects caused by diet composition on immune function parameters underline the importance of nutritional factors in the defense system of E. akaara.
Materials and Methods
Experimental design and sample preparation
This study was implemented at the Zhejiang province Key Lab of Mariculture and Enhancement in Zhoushan, China. Before the experiments, the juvenile fish were temporarily fed a commercial diet. At the beginning of the experiment, fish (initial weight 7.79 ± 0.01 g) were weighed and sorted into 9 cages (20 fish/cage). The experimental system consisted of 9 net cages (60 × 60 × 80 cm; L × W × H). All net cages were placed in one large concrete tank (13.0 × 4.0 × 1.5 m). A recirculating water system (including a sedimentation chamber, one drum filter, one fluidized sand filter, two biofilters, and two protein skimmers) was used during the experimental period. Three replicate groups of fish were used for each experiment diet. Three semi-purified isoproteic (48%) and isolipidic (9%) diets were formulated with different levels of corn starch (C1: 0%; C2: 18%; C3: 30%). All fish were fed to apparent satiation twice daily at 8:30 and 16:00, and the feeding trial lasted for 8 weeks.
During the experimental period, dissolved oxygen (DO), salinity, and temperature were 6.2 ± 0.1 mg/L, 25.1 ± 0.9 g/L and 27.5 ± 2.3 °C, respectively. At the end of the feeding trial, three fish in each cage were dissected, livers were immediately frozen in liquid nitrogen, and then they were stored at −80 °C until RNA extraction for sequencing. The experiment was implemented at the Zhejiang province Key Lab of Mariculture and Enhancement of Zhejiang Ocean University (Zhoushan, China). All experimental procedures were approved by the Local Animal Care and Use Committee (Zhejiang, China), and the study was carried out in accordance with regulations for the administration of affairs concerning experimental animals of China (Promulgated by Decree No. 2 of the State Science and Technology Commission on November 14, 1988).
RNA extraction and sequencing
Total RNA was extracted from livers of each group by homogenization in 1 ml TRIZol (Invitrogen) following the manufacturer’s instructions. The concentration and quality of total RNA were determined by electrophoresis (Thermo Scientific Nano drop 2000, USA). And total RNA was used for paired-end RNA sequencing. Library construction and sequencing were performed on an Illumina HiSeq. 2000 sequencer according to manufacturer’s specifications.
De novo assembly and functional annotation
Raw reads were pre-processed by discarding reads with adaptors, with unknown sequence (N) proportion greater than 5%, and those of low quality (Phred quality score <30). Then, raw reads were assembled by Trinity software using default parameters67. Transcripts shorter than 200 bp were removed from the subsequent analysis68. The assembly sequencing results evaluated used bowtie-1.0.169, which mapped the raw reads to the assembled transcriptome. Transcripts from the previous step were translated in all six possible open reading frames (ORFs) with TransDecoder tool, which was used to predict the ORF of the RNA sequencing assembly sequence. Proper translation was defined as one that gave the longest amino acid sequence70 according to the hidden Markov model (HMM).
The functional annotations of predicted amino acid sequences were performed using Trinotate (http://trinotate.sourceforge.net/) through searching against the Uniprot Knowledgebase and Swiss-Prot. In addition, we ran HMMER, signalIP, and tmHMM in Trinotate to identify protein domains, predict signal peptides, and transmembrane regions, respectively.
Gene Ontology (GO) annotations of transcripts were obtained by searching against the non-redundant (nr) database using the Blast2GO program71 with an E-value cutoff of 10−5. GO functional classifications were performed using WEGO software72. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were assigned to the assembled transcripts using the online KEGG Automatic Annotation Server (KAAS) (http://www.genome.jp/kegg/kaas/). The bi-directional best-hit (BBH) method was used to obtain KEGG orthology assignments.
Identification of differentially-expressed genes (DEGs) and RT-qPCR
DEGseq73 was used to compare RNA-seq data and identify differentially expressed genes. And the input of DEGseq is uniquely mapped reads from RNA-seq data with a gene annotation of the corresponding transcript expression values provided by RPKM74. A set of 20 genes which were significantly affected by high carbohydrate food were quantified by RT-qPCR to validate sequencing performance. Primers were designed using Primer premier 6 software (Table 4). The β-actin as the reference gene was quantified. For RT-qPCR, 2 μg of total RNA per sample was reverse transcribed into cDNA using the PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara), following manufacturer’s instructions. Amplifications were carried out in triplicate in a final volume of 20 μl containing 2 μl cDNA (1/10 dilution), 0.5 μM of each primer and 10 μl SYBR Green Supermix (Takara). The RT-qPCR profiles contained an initial activation step at 95 °C for 10 min, followed by 40 cycles: 15 s at 95 °C, 15 s at the specific primer pair annealing temperature (Ta; Table 4) and 15 s at 72 °C. After the amplification phase, a melt curve of 0.5 °C increments from 75 °C to 90 °C was performed, enabling confirmation of the amplification of a single product in each reaction. The analysis was based on the Ct values of the PCR products. Results are shown as changes in relative expression normalized with β-actin using the 2−ΔΔCt method. Chi-square statistic was applied to detect significant differences in each group.
Table 4.
Primers used for RT-qPCR analyses.
| Gene name | Primer sequence (5′–3′) | Fragment (bp) | Ta (°C) |
|---|---|---|---|
| TAP2 | TGGTTCGCAGCACAGTCA | 262 | 56 |
| GCCTCGTTATACCTCCTCAGT | |||
| Caspase 10 | GCTACGGACAAGACATAC | 257 | 55 |
| GGTGGATGATGAGGAGAA | |||
| CCL4 | TCTCGCTCTGTCTGTGTT | 251 | 56 |
| CGTCCAGGTAGGTGATGA | |||
| CD3 | TTCCAGTACCACAAGACAG | 159 | 53 |
| CCAGGACTCAGAGGTGTA | |||
| CIITA | GTCGGTTAGTCTGCTTGGT | 359 | 55 |
| TGTTCTGTCTGCTTCCTCTC | |||
| CXCL10 | TCTACCAAGCGACCATCT | 290 | 54 |
| GTGTCAGTGCTGTCAGTATA | |||
| CXCR4 | GACTCGGACTCTGTTGAC | 273 | 54 |
| CTGTGTTGGCATCTTCTTG | |||
| FOS | GGAGGTTGAGGTGTAGGT | 216 | 56 |
| GCTAATCTGTTGAAGGAGAAG | |||
| IKK1 | TTAACTCTTCTGGAACCTCTC | 263 | 55 |
| GTGACCTGACCGTATGGA | |||
| IL12 | CCTCACCATCTACATACACAT | 190 | 55 |
| CACCTTGACCTGGAACTG | |||
| IL8 | CCATCTGAGGAGAAGAACTC | 381 | 56 |
| CTGTGTTATTGAGCCTGATTC | |||
| IRF3 | GGAGTCGGCTTGAAGATA | 246 | 55 |
| TCAGTGTGGAAGAGGAAC | |||
| LCP2 | AGTCTGTCTGGCTGTGAT | 151 | 53 |
| TCTCCTCCTTCTTGCTGAT | |||
| NCF1 | TGGTGGTTCTGTCAGTGT | 376 | 58 |
| CCTTGTTGTGGATGCTCTT | |||
| NFATC | GTCTGCTTCATACTCGTCTAT | 136 | 53 |
| TGTCTGCCACTCTGTCAA | |||
| NFKBIA | TGTTGAGGAAGTCTGTGTT | 230 | 56 |
| GCTGAAGGAGGAGGAGTA | |||
| TLR2 | ACTCTGGAGGTGTTGGAT | 280 | 56 |
| CGTTGATGGCTGATTGGA | |||
| TLR9 | CCTACCTTGACATCTCTGAC | 282 | 52 |
| CCGCCTTACTGAAGAACAT | |||
| TNF12 | CTCGCCTCACATCTTCAG | 220 | 52 |
| TCCACAACAACTTCCACAA | |||
| TNFR11 | CAGTGCTGTCAGTCATCA | 127 | 52 |
| TGTATCCTCGTCCTCTTCA | |||
| β actin | CGACCTCACAGACTACCT | 221 | 56 |
| AACGGAACCTCTCATTGC |
Growth performance calculations
Data on initial body weight were used to calculate percentage weight gain (PWG). PWG (%) = 100% × (final body weight − initial body weight)/initial body weight. All measured values were presented as the mean ± standard deviation (SD). All data were tested for homogeneity (Levene’s test) and normal distribution (Kolmogorov-Smirnov tests), and necessary data were transformed before analysis. When data did not present variance homogeneity and normal distribution, Spearman’s correlation was used to determine the relationship between the response data and dietary carbohydrate. Values were regarded as statistically significant at P < 0.05. Statistical analysis was used to rank the groups using SPSS 18.0 (IBM, Chicago, USA) for Windows.
Acknowledgements
This work was supported by the Natural Science Foundation of Zhejiang Province of China [grant number LY16C190005); Public Technical Research and Social Development Program, Scientific and Technological Program of Zhoushan [grant number 2017C41008]; The Public Technical Research and Social Development Program, Scientific and Technological Program of Zhejiang [grant number 2015C33089]; The Open Foundation from Fishery Sciences in the First-Class Subjects of Zhejiang [grant number 20160011]; Scientific Research Foundation of Zhejiang Ocean University [grant number Q1418]; International Science and Technology Cooperation Program of Zhejiang Province [2015C34002] and Zhejiang Ocean University Scientific Research Initial Start Funding.
Author Contributions
J.W. and T.H. conceived the experiment, Y.Y. and X.L. conducted the experiments, Y.Y. and J.X. analysed the results. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bergot F, Breque J. Digestibility of starch by rainbow trout: effects of the physical state of starch and of the intake level. Aquaculture. 1983;34:203–212. doi: 10.1016/0044-8486(83)90203-X. [DOI] [Google Scholar]
- 2.Degani G, Viola S. The protein sparing effect of carbohydrates in the diet of eels (Anguilla anguilla) Aquaculture. 1987;64:283–91. doi: 10.1016/0044-8486(87)90191-8. [DOI] [Google Scholar]
- 3.Degani G, Viola S, Levanon D. Effects of dietary carbohydrate source on growth and body composition of the European eel (Anguilla anguilla L.) Aquaculture. 1986;52:97–104. doi: 10.1016/0044-8486(86)90029-3. [DOI] [Google Scholar]
- 4.Hemre, G.-I. Studies on carbohydrate nutrition in cod (Gadus morhua). University of Bergen, Norway: University of Bergen, Department of Fisheries and Marine Biology (2002).
- 5.Hemre G, Sandnes K, Waagbø R. Blood chemistry and organ nutrient composition in Atlantic salmon, Salmo salar L., fed graded amounts of wheat starch. Aquacult Nutr. 1995;1:37–42. doi: 10.1111/j.1365-2095.1995.tb00033.x. [DOI] [Google Scholar]
- 6.Rate JAG, Conversion F. and Body Composition of Catla catla, Labeo rohita, and Cirrhinus mrigala Fry Fed Diets of Various Carbohydrate-to-Lipid Ratios. J World Aquacult Soc. 1998;29:84–91. doi: 10.1111/j.1749-7345.1998.tb00303.x. [DOI] [Google Scholar]
- 7.Koops H, Tiews K, Tiews J, Gropp J. Stärke-und Fettverwertung netzkäfiggehaltener Regenbogenforellen (Salmo gairdneri) Aquaculture. 1974;4:277–286. doi: 10.1016/0044-8486(74)90040-4. [DOI] [Google Scholar]
- 8.Wilson R. Utilization of dietary carbohydrate by fish. Aquaculture. 1994;124:67–80. doi: 10.1016/0044-8486(94)90363-8. [DOI] [Google Scholar]
- 9.Furuichi, M. & Yone, Y. Change of blood sugar and plasma insulin levels of fishes [carp, Cyprinus carpio, red sea bream, Pagrus major, yellowtail, Seriola quinqueradiata] in glucose tolerance test. Bulletin of the Japanese Society of Scientific Fisheries (1981).
- 10.Ince BW, Thorpe A. Glucose and amino acid-stimulated insulin release in vivo in the European silver eel (Anguilla anguilla L.) Gen Comp Endocr. 1977;31:249–256. doi: 10.1016/0016-6480(77)90024-7. [DOI] [PubMed] [Google Scholar]
- 11.Palmer T, Ryman BE. Studies on oral glucose intolerance in fish. J Fish Biol. 1972;4:311–319. doi: 10.1111/j.1095-8649.1972.tb05680.x. [DOI] [Google Scholar]
- 12.Tashima L, Cahill G. Effects of insulin in the toadfish, Opsanus tau. Gen Comp Endocr. 1968;11:262–271. doi: 10.1016/0016-6480(68)90081-6. [DOI] [PubMed] [Google Scholar]
- 13.Thorpe A, Ince BW. The effects of pancreatic hormones, catecholamines, and glucose loading on blood metabolites in the northern pike (Esox lucius L.) Gen Comp Endocr. 1974;23:29–44. doi: 10.1016/0016-6480(74)90050-1. [DOI] [PubMed] [Google Scholar]
- 14.Wilson RP, Poe WE. Apparent inability of channel catfish to utilize dietary mono-and disaccharides as energy sources. J Nutr. 1987;117:280–285. doi: 10.1093/jn/117.2.280. [DOI] [PubMed] [Google Scholar]
- 15.Pieper A, Pfeffer E. Studies on the comparative efficiency of utilization of gross energy from some carbohydrates, proteins and fats by rainbow trout (Salmo gairdneri, R.) Aquaculture. 1980;20:323–332. doi: 10.1016/0044-8486(80)90093-9. [DOI] [Google Scholar]
- 16.Hemre GI, Mommsen T, Krogdahl Å. Carbohydrates in fish nutrition: effects on growth, glucose metabolism and hepatic enzymes. Aquacult Nutr. 2002;8:175–194. doi: 10.1046/j.1365-2095.2002.00200.x. [DOI] [Google Scholar]
- 17.Nemeth, E., Baird, A. W. & O’Farrelly, C. Microanatomy of the liver immune system. Seminars in immunopathology: Springer; 333–343 (2009). [DOI] [PubMed]
- 18.Castro R, et al. Early immune responses in rainbow trout liver upon viral hemorrhagic septicemia virus (VHSV) infection. PloS One. 2014;9:e111084. doi: 10.1371/journal.pone.0111084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golden-Mason L, O’Farrelly C. Having it all? Stem cells, haematopoiesis and lymphopoiesis in adult human liver. Immunol Cell Biol. 2002;80:45–51. doi: 10.1046/j.1440-1711.2002.01066.x. [DOI] [PubMed] [Google Scholar]
- 20.Ellis, A. Stress and the modulation of defence mechanisms in fish. Stress and fish (1981).
- 21.Maule A, Tripp R, Kaattari S, Schreck C. Stress alters immune function and disease resistance in chinook salmon (Oncorhynchus tshawytscha) J Endocr. 1989;120:135–142. doi: 10.1677/joe.0.1200135. [DOI] [PubMed] [Google Scholar]
- 22.Wiik R, Andersen K, Uglenes I, Egidius E. Cortisol-induced increase in susceptibility of Atlantic salmon, Salmo salar, to Vibrio salmonicida, together with effects on the blood cell pattern. Aquaculture. 1989;83:201–215. doi: 10.1016/0044-8486(89)90033-1. [DOI] [Google Scholar]
- 23.Braun WilliamA, Duvillard Serge,Pvon. Influence of carbohydrate delivery on the immune response during exercise and recovery from exercise. Nutrition. 2004;20:645–650. doi: 10.1016/j.nut.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 24.Cowey C, Adron J, Brown D, Shanks AM. Studies on the nutrition of marine flatfish. The metabolism of glucose by plaice (Pleuronectes platessa) and the effect of dietary energy source on protein utilization in plaice. Brit J Nutr. 1975;33:219–231. doi: 10.1079/BJN19750026. [DOI] [PubMed] [Google Scholar]
- 25.Ellis SC, Reigh RC. Effects of dietary lipid and carbohydrate levels on growth and body composition of juvenile red drum. Sciaenops ocellatus. Aquaculture. 1991;97:383–394. doi: 10.1016/0044-8486(91)90330-A. [DOI] [Google Scholar]
- 26.Hilton J, Atkinson J. Response of rainbow trout (Salmo gairdneri) to increased levels of available carbohydrate in practical trout diets. Brit J Nutr. 1982;47:597–607. doi: 10.1079/BJN19820071. [DOI] [PubMed] [Google Scholar]
- 27.Mohapatra M, Sahu N, Chaudhari A. Utilization of gelatinized carbohydrate in diets of Labeo rohita fry. Aquacult Nutr. 2003;9:189–196. doi: 10.1046/j.1365-2095.2003.00243.x. [DOI] [Google Scholar]
- 28.Mohanta KN, Mohanty SN, Jena J, Sahu NP, Patro B. Carbohydrate level in the diet of silver barb, Puntius gonionotus (Bleeker) fingerlings: effect on growth, nutrient utilization and whole body composition. Aquac Res. 2009;40:927–937. doi: 10.1111/j.1365-2109.2009.02186.x. [DOI] [Google Scholar]
- 29.Li XF, Wang Y, Liu WB, Jiang GZ, Zhu J. Effects of dietary carbohydrate/lipid ratios on growth performance, body composition and glucose metabolism of fingerling blunt snout bream Megalobrama amblycephala. Aquacult Nutr. 2013;19:701–708. doi: 10.1111/anu.12017. [DOI] [Google Scholar]
- 30.Ren M, Ai Q, Mai K, Ma H, Wang X. Effect of dietary carbohydrate level on growth performance, body composition, apparent digestibility coefficient and digestive enzyme activities of juvenile cobia, Rachycentron canadum L. Aquac Res. 2011;42:1467–1475. doi: 10.1111/j.1365-2109.2010.02739.x. [DOI] [Google Scholar]
- 31.Wang J, et al. Dietary protein requirement of juvenile red spotted grouper (Epinephelus akaara) Aquaculture. 2016;450:289–294. doi: 10.1016/j.aquaculture.2015.08.007. [DOI] [Google Scholar]
- 32.Jiang YD, Wang JT, Han T, Li XY, Hu SX. Effect of dietary lipid level on growth performance, feed utilization and body composition by juvenile red spotted grouper (Epinephelus akaara) Aquac Int. 2015;23:99–110. doi: 10.1007/s10499-014-9801-7. [DOI] [Google Scholar]
- 33.Krogdahl Å, Hemre GI, Mommsen T. Carbohydrates in fish nutrition: digestion and absorption in postlarval stages. Aquacult Nutr. 2005;11:103–122. doi: 10.1111/j.1365-2095.2004.00327.x. [DOI] [Google Scholar]
- 34.Comfort, A. Ageing. The biology of senescence. Ageing The biology of senescence (1964).
- 35.DeChiara, T. M., Efstratiadis, A. & Robertsen, E. J. A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting (1990). [DOI] [PubMed]
- 36.Baker J, Liu J-P, Robertson EJ, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. doi: 10.1016/S0092-8674(05)80085-6. [DOI] [PubMed] [Google Scholar]
- 37.Moriyama S, et al. Development of a homologous radioimmunoassay for coho salmon insulin-like growth factor-I. Gen Comp Endocr. 1994;96:149–161. doi: 10.1006/gcen.1994.1167. [DOI] [PubMed] [Google Scholar]
- 38.BAIL PY, et al. Structure, Function, and Regulation of Insulin-like Growth Factors in Fish. Ann NY Acad Sci. 1998;839:157–161. doi: 10.1111/j.1749-6632.1998.tb10750.x. [DOI] [PubMed] [Google Scholar]
- 39.Reindl KM, Sheridan MA. Peripheral regulation of the growth hormone-insulin-like growth factor system in fish and other vertebrates. Comp Biochem Phys A. 2012;163:231–245. doi: 10.1016/j.cbpa.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 40.Wood AW, Duan C, Bern HA. Insulin-like growth factor signaling in fish. Int Rev Cytol. 2005;243:215–285. doi: 10.1016/S0074-7696(05)43004-1. [DOI] [PubMed] [Google Scholar]
- 41.Côté G, Perry G, Blier P, Bernatchez L. The influence of gene-environment interactions on GHR and IGF-1 expression and their association with growth in brook charr, Salvelinus fontinalis (Mitchill) BMC genetics. 2007;8:87. doi: 10.1186/1471-2156-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Safari, R., Hoseinifar, S. H., Nejadmoghadam, S. & Khalili, M. Non-specific immune parameters, immune, antioxidant and growth-related genes expression of common carp (Cyprinus carpio L.) fed sodium propionate. Aquac Res (2017).
- 43.Hoseinifar SH, Safari R, Dadar M. Dietary sodium propionate affects mucosal immune parameters, growth and appetite related genes expression: Insights from zebrafish model. Gen Comp Endocr. 2017;243:78–83. doi: 10.1016/j.ygcen.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 44.Kelley KM, Siharath K, Bern HA. Identification of insulin‐like growth factor‐binding proteins in the circulation of four teleost fish species. J Exp Zool. 1992;263:220–224. doi: 10.1002/jez.1402630213. [DOI] [PubMed] [Google Scholar]
- 45.Duan C, Xu Q. Roles of insulin-like growth factor (IGF) binding proteins in regulating IGF actions. Gen Comp Endocr. 2005;142:44–52. doi: 10.1016/j.ygcen.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 46.Calduch-Giner JA, Mingarro M, de Celis SV-Rn, Boujard D, Pérez-Sánchez J. Molecular cloning and characterization of gilthead sea bream (Sparus aurata) growth hormone receptor (GHR). Assessment of alternative splicing. Comp Biochem Phys B. 2003;136:1–13. doi: 10.1016/S1096-4959(03)00150-7. [DOI] [PubMed] [Google Scholar]
- 47.Uchida K, et al. Effects of fasting on growth hormone/insulin-like growth factor I axis in the tilapia, Oreochromis mossambicus. Comp Biochem Phys A. 2003;134:429–439. doi: 10.1016/S1095-6433(02)00318-5. [DOI] [PubMed] [Google Scholar]
- 48.Beckman BR, Shimizu M, Gadberry BA, Cooper KA. Response of the somatotropic axis of juvenile coho salmon to alterations in plane of nutrition with an analysis of the relationships among growth rate and circulating IGF-I and 41kDa IGFBP. Gen Comp Endocr. 2004;135:334–344. doi: 10.1016/j.ygcen.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 49.Duan C. The insulin-like growth factor system and its biological actions in fish. Am Zool. 1997;37:491–503. doi: 10.1093/icb/37.6.491. [DOI] [Google Scholar]
- 50.Reinecke M, et al. Growth hormone and insulin-like growth factors in fish: where we are and where to go. Gen Comp Endocr. 2005;142:20–24. doi: 10.1016/j.ygcen.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 51.Shimizu M, et al. Circulating salmon 28-and 22-kDa insulin-like growth factor binding proteins (IGFBPs) are co-orthologs of IGFBP-1. Gen Comp Endocr. 2011;174:97–106. doi: 10.1016/j.ygcen.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 52.Shimizu M, Suzuki S, Horikoshi M, Hara A, Dickhoff WW. Circulating salmon 41-kDa insulin-like growth factor binding protein (IGFBP) is not IGFBP-3 but an IGFBP-2 subtype. Gen Comp Endocr. 2011;171:326–331. doi: 10.1016/j.ygcen.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 53.Macqueen, D. J., Johnston, I. A. Evolution of ancient functions in the vertebrate insulin-like growth factor system uncovered by study of duplicated salmonid fish genomes. Mol Biol Evol mst017 (2013). [DOI] [PMC free article] [PubMed]
- 54.Alvarez-Pellitero P. Fish immunity and parasite infections: from innate immunity to immunoprophylactic prospects. Vet Immunol Immunop. 2008;126:171–198. doi: 10.1016/j.vetimm.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 55.Secombes C, et al. Cytokines and innate immunity of fish. Dev Comp Immunol. 2001;25:713–723. doi: 10.1016/S0145-305X(01)00032-5. [DOI] [PubMed] [Google Scholar]
- 56.Cartier L, Hartley O, Dubois-Dauphin M, Krause K-H. Chemokine receptors in the central nervous system: role in brain inflammation and neurodegenerative diseases. Brain Res Rev. 2005;48:16–42. doi: 10.1016/j.brainresrev.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 57.Kindt, T. J., Goldsby, R. A., Osborne, B. A. & Kuby, J. Kuby immunology: Macmillan (2007).
- 58.Offermanns, S. Encyclopedia of molecular pharmacology: Springer Science & Business Media (2008).
- 59.Tanekhy M. The role of Toll‐like Receptors in innate immunity and infectious diseases of teleost. Aquac Res (2014).
- 60.Bakke-McKellep AM, et al. Response to soy: T-cell-like reactivity in the intestine of Atlantic salmon, Salmo salar L. J Fish Dis. 2007;30:13e25. doi: 10.1111/j.1365-2761.2007.00769.x. [DOI] [PubMed] [Google Scholar]
- 61.Bakke-McKellep AM, et al. Effects of dietary soyabean meal, inulin and oxytetracycline on intestinal microbiota and epithelial cell stress, apoptosis and proliferation in the teleost Atlantic salmon (Salmo salar L.) Br J Nutr. 2007;97:699e713. doi: 10.1017/S0007114507381397. [DOI] [PubMed] [Google Scholar]
- 62.Krogdahl Å, Bakke-McKellep AM, Røed KH, Baeverfjord G. Feeding Atlantic salmon Salmo salar L. soybean products: effects on disease resistance (furunculosis), and lysozyme and IgM levels in the intestinal mucosa. Aquacult Nutr. 2000;6:77e84. doi: 10.1046/j.1365-2095.2000.00129.x. [DOI] [Google Scholar]
- 63.Krogdahl Å, Bakke-McKellep AM, Baeverfjord G. Effects of graded levels of standard soybean meal on intestinal structure, mucosal enzyme activities, and pancreatic response in Atlantic salmon (Salmo salar L.) Aquacult Nutr. 2003;9:361e71. doi: 10.1046/j.1365-2095.2003.00264.x. [DOI] [Google Scholar]
- 64.Lilleeng E, Frøystad MK, Østby GC, Valen EC, Krogdahl Å. Effects of diets containing soybean meal on trypsin mRNA expression and activity in Atlantic salmon (Salmo salar L) Comp Biochem Physiol A Mol Integr Physiol. 2007;147:25e36. doi: 10.1016/j.cbpa.2006.10.043. [DOI] [PubMed] [Google Scholar]
- 65.Skugor S, et al. Gene expression responses to restricted feeding and extracted soybean meal in Atlantic salmon (Salmo salar L.) Aquacult Nutr. 2011;17:505e17. doi: 10.1111/j.1365-2095.2010.00832.x. [DOI] [Google Scholar]
- 66.Sahlmann C, Sutherland BJ, Kortner TM, et al. Early response of gene expression in the distal intestine of Atlantic salmon (Salmo salar L.) during the development of soybean meal induced enteritis. Fish Shellfish Immu. 2013;34:599–609. doi: 10.1016/j.fsi.2012.11.031. [DOI] [PubMed] [Google Scholar]
- 67.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saha S, et al. Using the transcriptome to annotate the genome. Nat Biotechnol. 2002;20:508–512. doi: 10.1038/nbt0502-508. [DOI] [PubMed] [Google Scholar]
- 71.Conesa A, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 72.Ye J, et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:293–297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biol. 2010;11:106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]