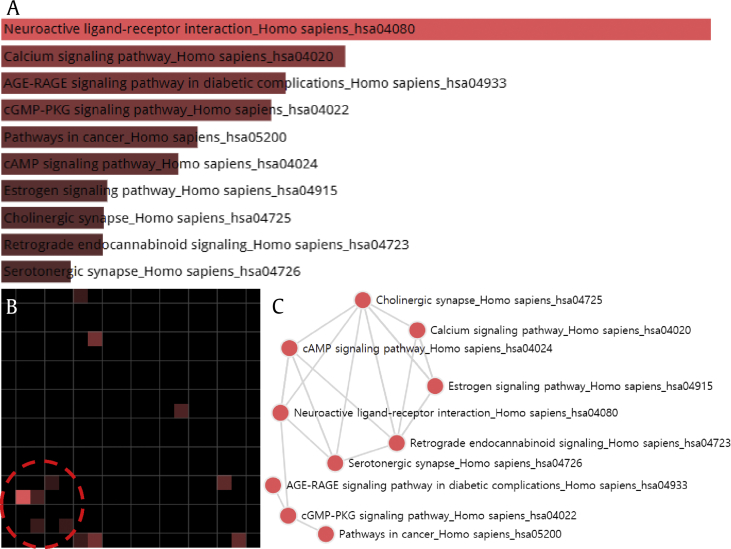
Fig. 6.
Grid and network analysis of enriched terms using the KEGG 2016 library. The gene symbols of 161 targets are used as an input and the result is displayed in three different manners: a bar graph, a grid, and a network. (A) The top 10 terms are ranked as bar graph in descending order. The length of the bar represents the significance of the specific gene-sets represented by the terms. The brighter the color, the more significant that term is. (B) Each grid square represents a term and is arranged based on term-term similarity, which represents one term's gene-set content similarity with another term. It shows the top 10 terms sorted by enrichment score. The brighter the square, the more significant that term is. A circle is used to highlight the most dominant cluster of enriched terms: neuroactive ligand–receptor interaction, cAMP signaling pathway, serotonergic synapse, cholinergic synapse, and retrograde endocannabinoid signaling (displayed clockwise from the brightest grid). (C). In the network, each node represents the enriched term, and the edge between two nodes means that the two terms have some gene content similarity.