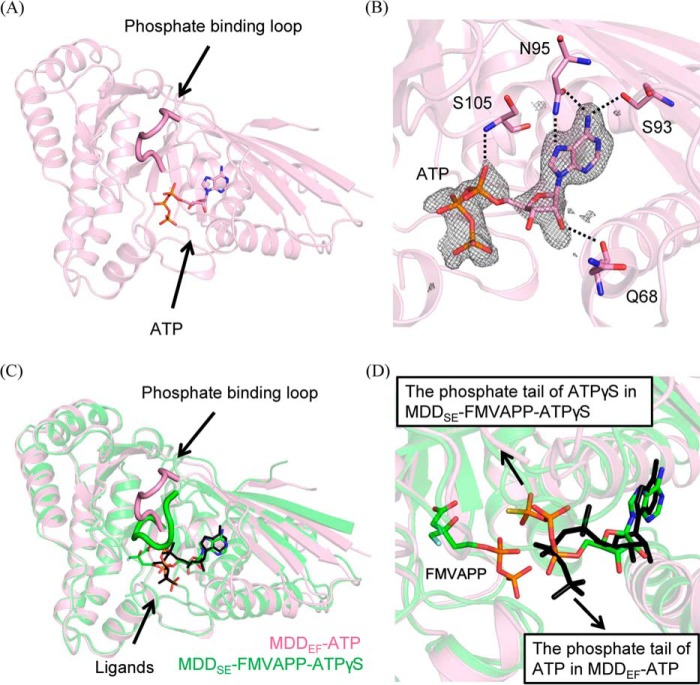
Figure 3.
Superimposition of complex structures of MDDEF–ATP and MDD from S. epidermidis bound with FMVAPP and ATPγS (MDDS.E.–FMVAPP–ATPγS). A, ribbon model of the crystal structure of MDDEF–ATP is shown in pink with the bound ATP molecule as a stick model. B, ATP molecule is surrounded by the SA-omit map (mFo − DFc at a contour of 3σ, cropped at 5 Å from ATP). Residues (Gln-68, Ser-93, Asn-95, and Ser-105) that form hydrogen bonds with ATP are shown as stick models. The hydrogen bonding partners (Gln-68–O and ATP–O2′, 3.3 Å; Ser-93–Oγ and ATP–N6, 3.0 Å; Asn-95–Oδ and ATP–N6, 2.9 Å; Asn-95–Nδ and ATP–N7, 3.3 Å; Ser-105–N and ATP–Oα, 2.9 Å) are connected by dashed lines. C, overlay of the models of MDDSE–FMVAPP–ATPγS (PDB code 4DPT, green) and MDDEF–ATP (pink) (r.m.s.d. = 0.66 Å) is depicted with the phosphate-binding loops emphasized. D, ligands from two structures are shown as stick models (FMVAPP and ATPγS from MDDSE–FMVAPP–ATPγS; ATP from MDDEF–ATP). The arrows in black indicate the distinct orientations of the phosphate tails of ATPγS and ATP from these two structures.