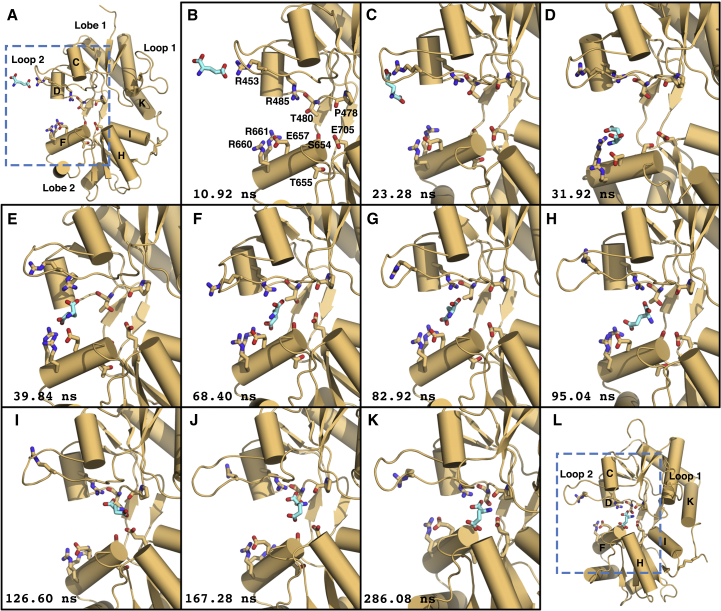
Figure 1.
Dynamics of Glutamate Binding
Time points (lower left of each panel) are relative to the start of Movie S1. This trajectory corresponds to the LBD dimer system Tdim1 (see Table S1); only the LBD that binds glutamate is shown for clarity.
(A) Prior to ligand entry to the binding pocket, the LBD is open; (ξ1, ξ2) = (12.3, 12.2 Å). The ligand’s γ-carboxylate contacts R453 on Lobe 1.
(B) Close-up view of (A).
(C) Glutamate slips into the binding cleft.
(D) The ligand contacts R661 on Lobe 2.
(E) A metastable interaction forms across Lobes 1 and 2. The ligand’s γ-carboxylate contacts E657, R660, and R661 on helix F, and the ligand’s α-carboxylate contacts R485. R485 flickers out of the binding pocket to interact with the ligand.
(F) R485 relaxes toward the binding pocket.
(G) The metastable interaction at the ligand’s α- and γ-carboxylate between Lobes 1 and 2 persists.
(H) The ligand's α-carboxylate and amine move to interact with binding pocket residues in Lobe 1.
(I) The ligand shifts into the binding pocket, with its α-carboxylate contacting R485. Lobe 2 interactions with helix F are broken. In the pocket, the ligand’s amine is coordinated by P478 and L480. Cleft closure is initiated once helix F undergoes a backward tilt to form a pocket for the ligand’s γ-carboxylate.
(J) Glutamate adopts the crystallographic conformation.
(K) As the cleft closes to secure the ligand, the ligand’s amine contacts E705 on Lobe 2, and the ligand’s γ-carboxylate contacts the backbone amine of S654.
(L) Expanded view of (K). The LBD closes around the ligand in the crystallographic conformation: (ξ1, ξ2) = (11.8, 10.8 Å).
See also Figures S1 and S8, Movie S1, and Table S1.