Figure 2.
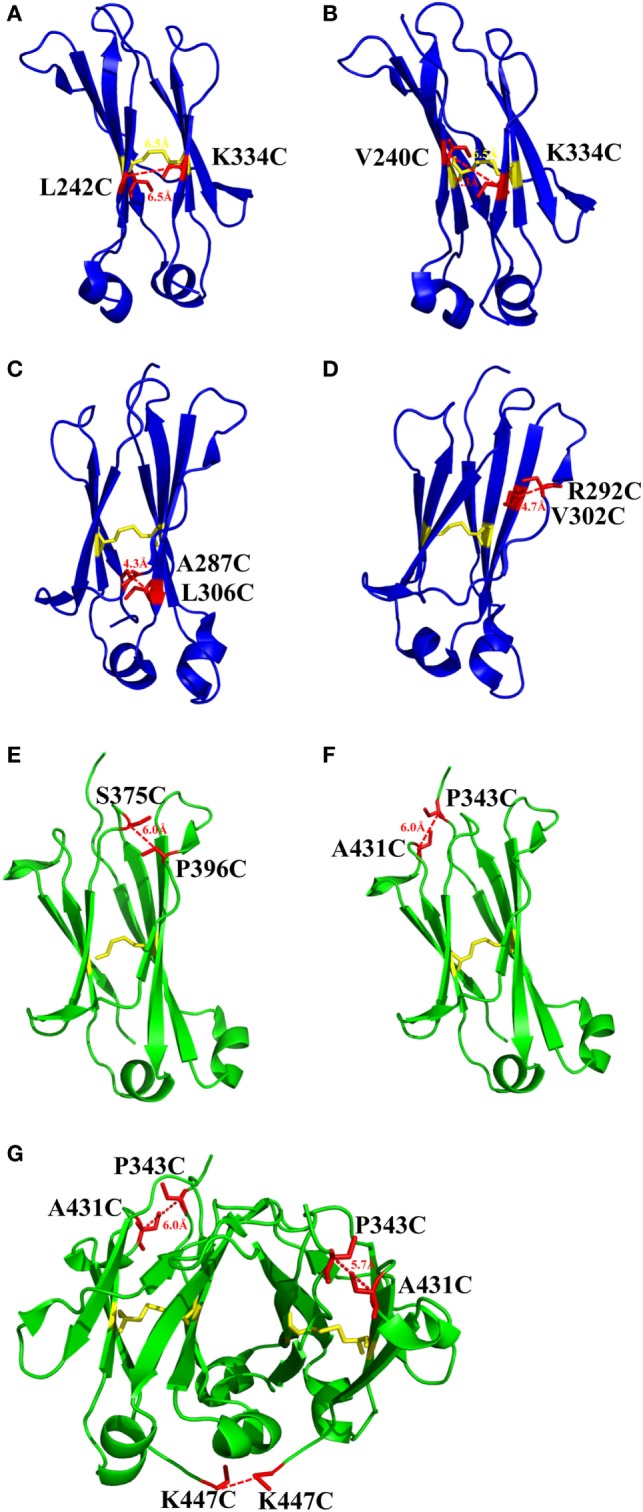
The structures of CH2 and CH3 domains [PDB 3AVE (47)] with different mutations for introduction of additional disulfide bonds presented by PyMOL. (A) CH2 mutant L242C/K334C. (B) CH2 mutant V240C/K334C. (C) CH2 mutant A287C/L306C. (D) CH2 mutant R292C/V302C. (E) CH3 mutant S375C/P396C. (F) CH3 mutant P343C/A431C. (G) CH3 mutant P343C/A431C/K447C. The native disulfide bond is colored by yellow, whereas the mutated residues for additional disulfide bonds are colored by red. All the marked distance is the measured between two α-carbon atoms in related two cysteines after mutagenesis by using PyMOL.
