Figure 3.
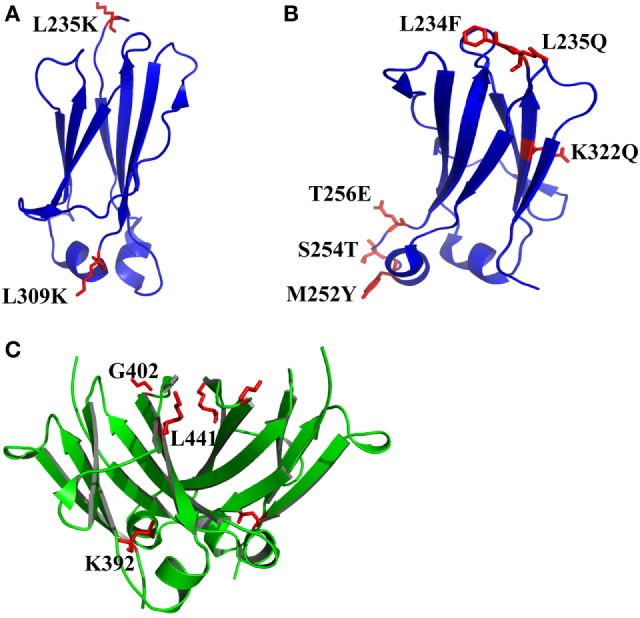
The structures of CH2 and CH3 domains [PDB 3AVE (47)] with optimized non-covalent interactions presented by PyMOL. (A) CH2 L235K/L309K mutant. (B) CH2 L234F/L235Q/K322Q/M252Y/S254T/T256E mutant (FQQ–YTE). (C) The residues K392, G402, and L441 in human IgG1 CH3 domain, which can be used to replace the corresponding residues in the bovine CH3 domain (G197, S207, and T246) for increase of the stability. The mutated residues for additional disulfide bonds are colored by red.
