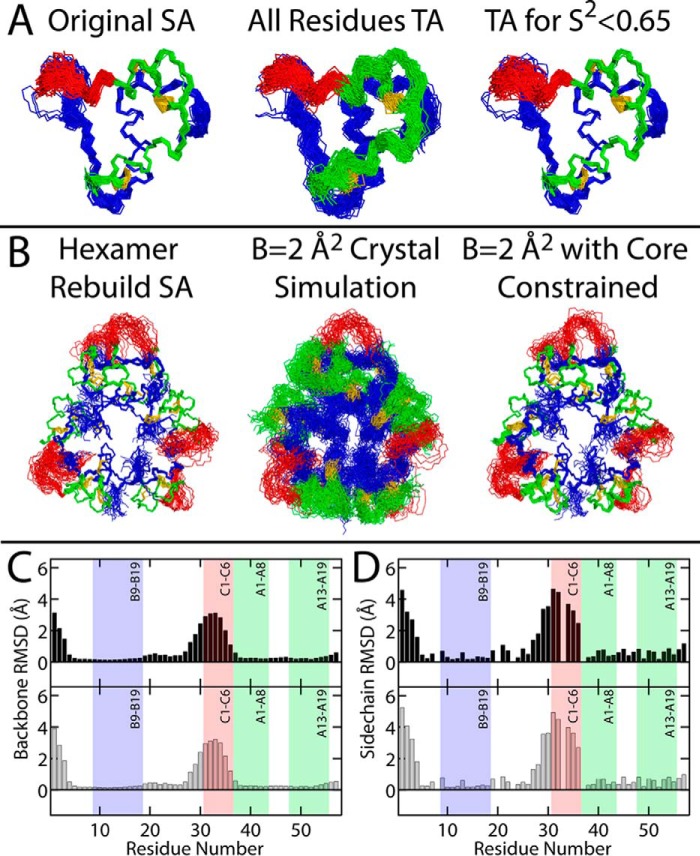
Figure 9.
Alternative structural ensembles of SCI-b and SCI-a. A, time-averaged NOE-based ensemble of 98 structures of SCI-b using a standard SA protocol (original SA; left). Each structure was run through a TA distance-restrained MD simulation wherein MD time-averaging was activated for all restraints (All Residues TA; middle). A separate simulation was then performed that enforced time-averaged restraints only for residues residing within flexible regions (predicted S2 < 0.65; right). B, each of the 35 hexameric structures generated using carbon–carbon distance restraints derived from the single-structure SCI-a crystallographic refinement (Hexamer Rebuild SA; left) were subjected to multiconformer simulation (see Fig. S6A for schematic description of the simulation procedure) with all thermal B-factors set to 2 Å2 and either no residues constrained (B = 2 Å2 crystal simulation; middle) or with the positions of B5–B26 and A1–A21 constrained (B = 2 Å2 with core constrained; right). The main-chain (C) and heavy-atom side-chain (D) r.m.s.d. per residue for SCI-b (black) or SCI-a (gray) were calculated from ensembles in A (right) and B (right), respectively. The r.m.s.d. for the SCI-a hexamer ensemble are averaged over all monomers. The B9–B19, A1–A8, and A13–A19 helices and C1–C6 segment are shown as shaded boxes. Gly residues at positions B8, B20, B23, C3, and A1 were excluded from heavy-atom side-chain r.m.s.d. calculations.