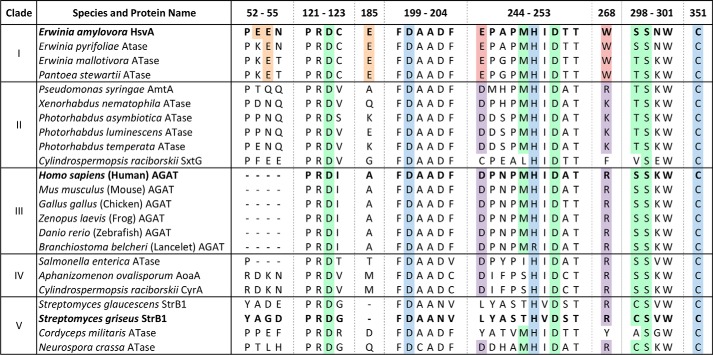
Figure 7.
Amino acid sequence alignment of selected regions from known or predicted amidinotransferases. Selected regions from the 23 amino acid sequences used in the phylogram depicted in Fig. 1 were aligned using Clustal Omega and annotated manually (the full sequence alignment is given in Fig. S1). The clade designations from the phylogram in Fig. 1 are listed on the left. The UniProt accession numbers for the aligned sequences may be found in Table S1. The regions are numbered according to the amino acid positions in the HsvA sequence. The amidinotransferase proteins for which the molecular structure is known (HsvA, hAGAT, and StrB1) are highlighted in boldface type. Highlighted in blue are the strictly conserved amino acids forming the catalytic triad (Asp200, His249, and Cys351 in HsvA). Conserved amino acids outside of the catalytic triad that interact directly with the arginine substrate in hAGAT are highlighted in green (7). Highlighted in red are the Glu and Trp residues located at the opening of the substrate access channel in HsvA, along with conserved amino acids found in other homologs in clade I. The corresponding amino acids in the same positions in homologs from other clades are highlighted in purple. Glu residues on the surface of HsvA near the opening of the substrate access channel are highlighted in orange, along with conserved amino acids at corresponding positions in other homologs in clade I.