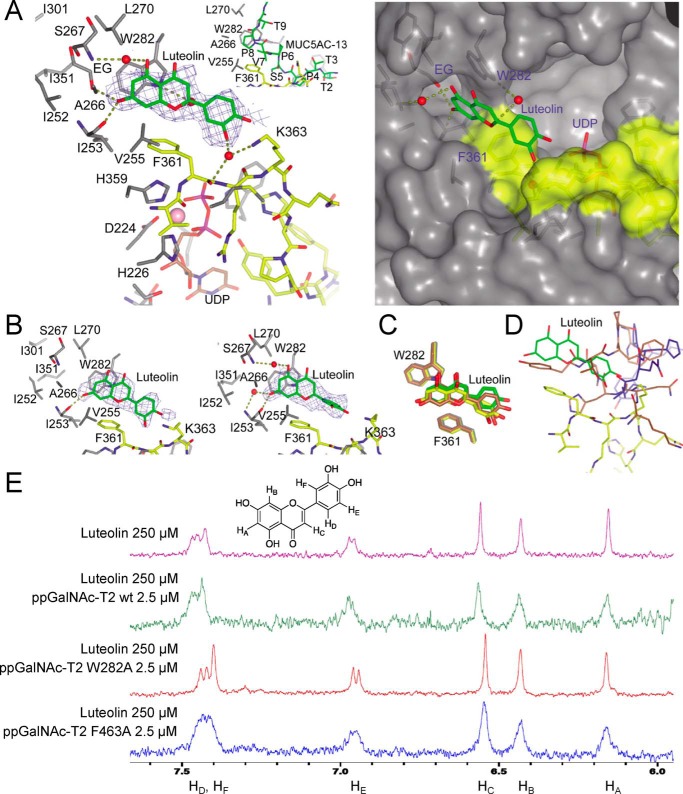
Figure 3.
X-ray structure and 1H NMR binding experiments of ppGalNAc-T2/mutants with luteolin. A, view (left) of the luteolin-binding site of the ppGalNAc-T2·UDP-luteolin complex. The crystal structure of ppGalNAc-T2 in complex with UDP and MUC5AC-13 (32) is shown for comparison purposes (inset). A surface representation (right) of the ppGalNAc-T2·UDP-luteolin complex depicts the pocket present in these enzymes. Part of the residues forming the luteolin-binding site and the pocket are shown in gray, whereas residues of the flexible loop are depicted in yellow. Luteolin/MUC5AC-13 and UDP are shown as green and brown carbon atoms, respectively. Ethylene glycol is shown as gray carbon atoms, whereas water molecules are depicted as red spheres. Hydrogen bond interactions are shown as dotted deep olive lines. Electron density maps are Fo − Fc syntheses (blue) contoured at 2.2 σ for luteolin. B, view of the two additional luteolin molecules present in the AU. C, overlay of the ppGalNAc-T2 molecules in complex with luteolin showing the flexibility mainly in the dihydroxyphenyl moiety. Luteolin and Trp282/Phe361 are depicted as green, yellow, and brown carbon atoms. D, overlay of one of the monomers containing luteolin with ppGalNAc-T2 inactive form in complex with UDP (Protein Data Bank entry 2FFV (31)) and ppGalNAc-T2 inactive form in complex with MUC5AC-Cys13 (Protein Data Bank entry 5AJN (32)). This figure shows that luteolin has steric hindrance with both inactive forms of ppGalNAc-T2. The flexible loop for the active and inactive forms is shown in yellow and blue/brown, respectively. E, 1H NMR spectra were recorded to compare the effect of binding on the luteolin signal broadening. Shown are 1H NMR spectra of free luteolin and luteolin with ppGalNAc-T2 wild type and the mutants W282A and F463A.